library(tidyCoverage)
library(tidySummarizedExperiment)
library(rtracklayer)
library(plyranges)
library(purrr)
library(ggplot2)
# ~~~~~~~~~~~~~~~ Import genomic features into a named list ~~~~~~~~~~~~~~~ #
features <- list(
TSSs = system.file("extdata", "TSSs.bed", package = "tidyCoverage"),
conv_sites = system.file("extdata", "conv_transcription_loci.bed", package = "tidyCoverage")
) |> map(~ import(.x))
# ~~~~~~~~~~~~ Import coverage tracks into a `BigWigFileList` ~~~~~~~~~~~~~ #
tracks <- list(
Scc1 = system.file("extdata", "Scc1.bw", package = "tidyCoverage"),
RNA_fwd = system.file("extdata", "RNA.fwd.bw", package = "tidyCoverage"),
RNA_rev = system.file("extdata", "RNA.rev.bw", package = "tidyCoverage"),
PolII = system.file("extdata", "PolII.bw", package = "tidyCoverage"),
MNase = system.file("extdata", "MNase.bw", package = "tidyCoverage")
) |> BigWigFileList()CE <- CoverageExperiment(tracks, features, width = 1000, ignore.strand = FALSE)
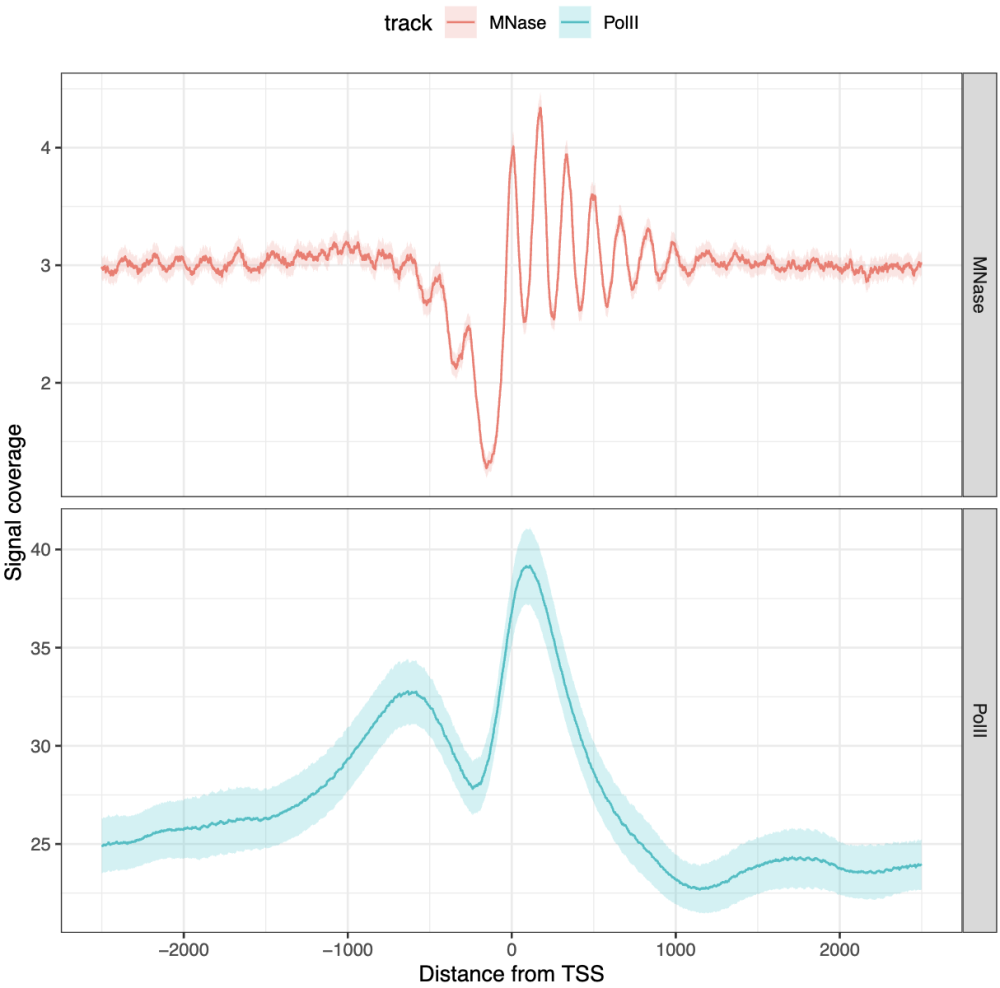
CE |>
filter(track %in% c('MNase', 'PolII')) |>
filter(features == 'TSSs') |>
aggregate() |>
ggplot(aes(x = coord, y = mean)) +
geom_ribbon(aes(ymin = ci_low, ymax = ci_high, fill = track), alpha = 0.2) +
geom_line(aes(col = track)) +
facet_grid(track ~ ., scales = "free") +
labs(x = 'Distance from TSS', y = 'Signal coverage') +
theme_bw() +
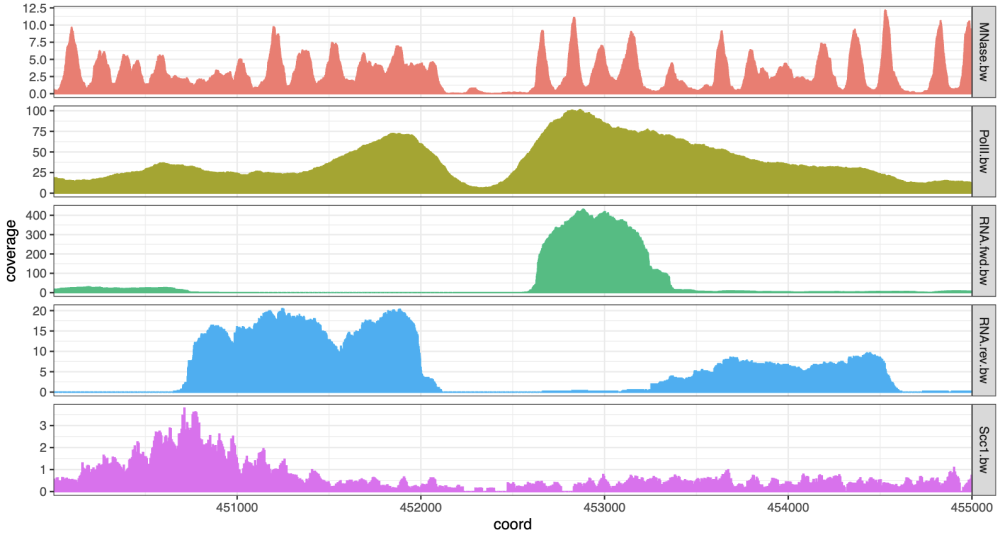
theme(legend.position = 'top')CoverageExperiment(tracks, GRanges("II:450001-455000"), width = 5000) |>
expand() |>
ggplot(aes(x = coord, y = coverage)) +
geom_col(aes(fill = track, col = track)) +
facet_grid(track~., scales = 'free') +
scale_x_continuous(expand = c(0, 0)) +
theme_bw() +
theme(legend.position = "none", aspect.ratio = 0.1)A number of CRAN, Bioconductor or GitHub packages already exist to enable genomic track
data visualization, for instance:
Gviz[Bioconductor]soGGi[Bioconductor]GenomicPlot[Bioconductor]plotgardener[Bioconductor]genomation[Bioconductor]ggcoverage[GitHub]GenomicScores[Bioconductor]
Compared to these existing solutions, tidyCoverage directly extends SummarizedExperiment infrastructure and
follows tidy "omics" principles. It does
not directly provide plotting functionalities, but instead focuses on data recovery, structure and coercion,
using a familiar grammar and standard representation of the data.
This ensures seamless integration of genomic track investigation in exisiting
Bioconductor and data analysis workflows.