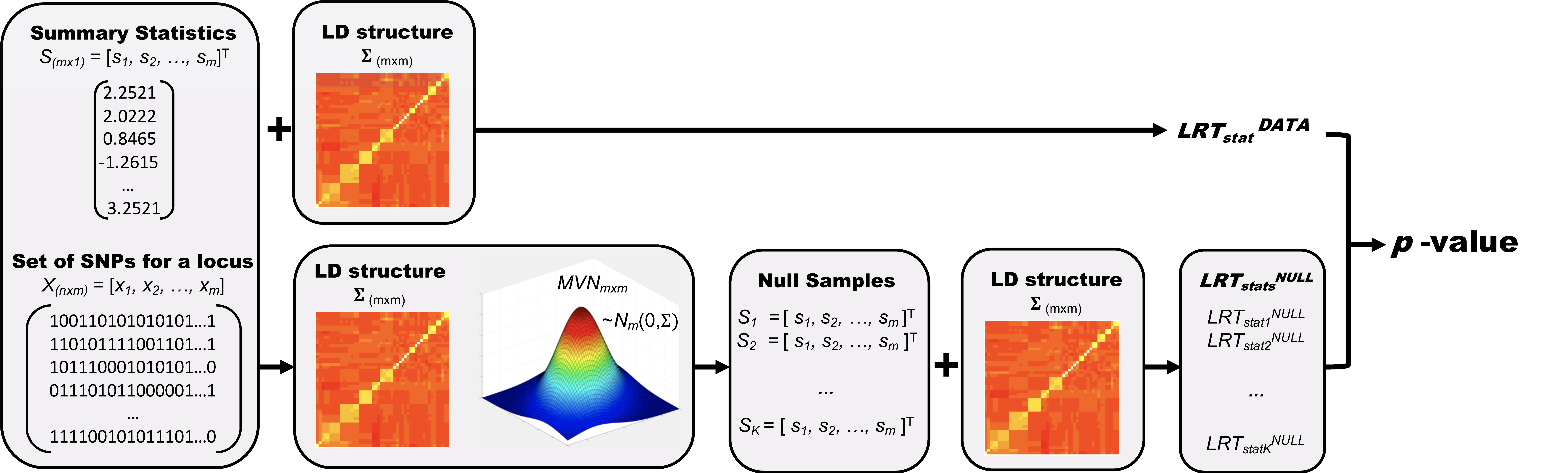
In standard genome-wide association studies (GWAS), the standard association test is underpowered to detect associations between loci with multiple causal variants with small effect sizes. We propose a statistical method, Model-based Association test Reflecting causal Status (MARS), that finds associations between variants in risk loci and a phenotype, considering the causal status of variants, only requiring the existing summary statistics to detect associated risk loci. Utilizing extensive simulated data and real data, we show that MARS increases the power of detecting true associated risk loci compared to previous approaches that consider multiple variants, while controlling the type I error.
MARS: leveraging allelic heterogeneity to increase power of association testing. Genome Biol 22, 128 (2021). [Links]
- The C++ library for GNU GSL is required.
- makesigmasemiPDRcppGSL is required.
- marsR works only on *nix (Linux, Unix such as macOS) system. please check .Platform$OS.type function.
- We currently only support R 3.5+.*
install.packages("data.table")
install.packages("Rcpp")
install.packages("RcppGSL")
install.packages("RcppArmadillo")
install.packages("devtools")
devtools::install_github("junghyunJJ/makesigmasemiPDRcppGSL")
devtools::install_github("junghyunJJ/marsR")
library(marsR)
> data(testdata)
> mars(testdata$stat, testdata$geno, threshold = 5e-08)
[2020-04-07 09:08:40] - calculating MARS alt LRT
[2020-04-07 09:08:40] - make null data
[2020-04-07 09:08:40] - calculating MARS null LRT
|++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=10s
Total analysis time: 10.97 secs
$alt
LRT uni
1 15985.79 1.968e-07
$results
LRT_pvalue univariate_pvalue threshold_pvalue threshold_UNI significance_LRT significance_UNI
1 0 0.00099551 0.00099551 5e-08 TRUE FALSE