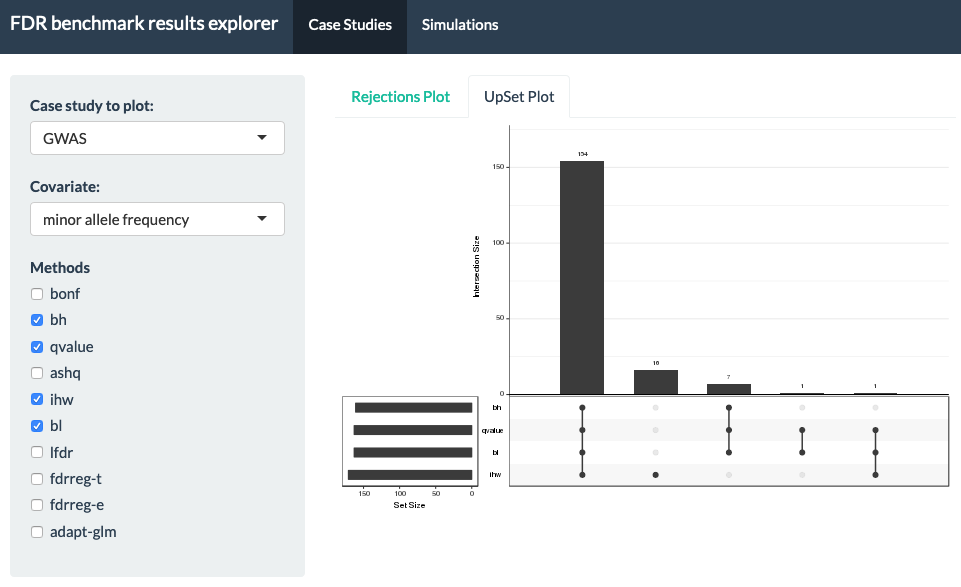
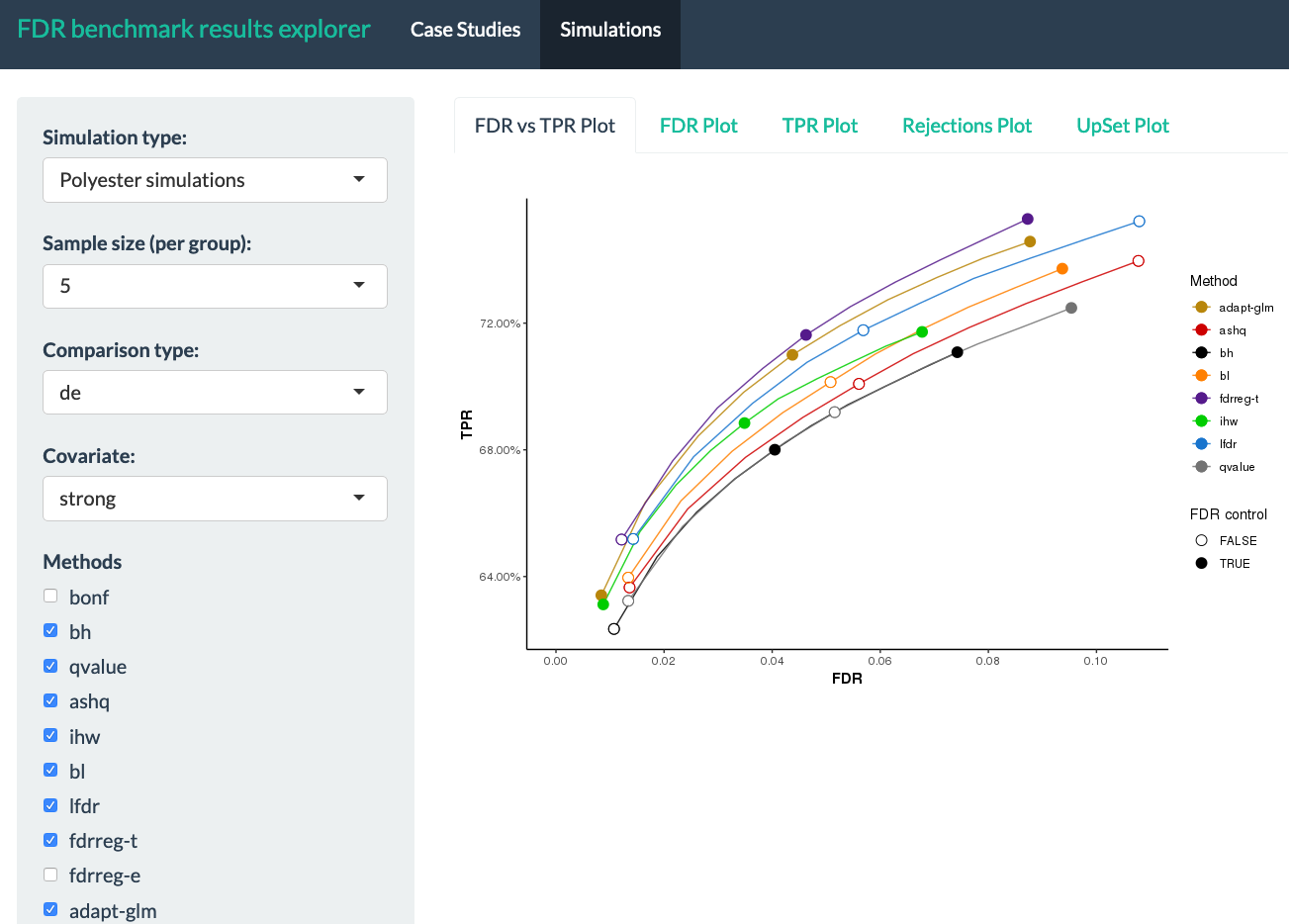
This is a Shiny App for exploring results from "A practical guide to methods controlling false discoveries in computational biology" by Korthauer, Kimes et al. (2019). In this paper, we generated results from a series of case studies of high-throughput biology, as well as extensive simulation using both resampling techniques and direct simulation of RNA-seq counts using Polyester. This tool enables interactive exploration of these results.
The tool provides evaluation plots for case studies:
- number of rejections by specified alpha
- overlap among sets using UpSet plots
For the yeast in silico resampling experiments and Polyester simulation studies (over 100 replications):
- mean FDR versus TPR
- FDR by specified alpha
- TPR by specified alpha
- number of rejections by specified alpha
- average overlap among sets using UpSet plots
For each of these plots, you can toggle between different selections of:
- informative covariates
- datasets
- simulation settings (e.g. strength of covariate, modality of alternative, sample size, proportion of null hypotheses)
- methods (e.g. subset to compare just Benjamini-Hochberg with IHW)
install.packages("BiocManager")
install.packages("devtools")
BiocManager::install(shiny)
BiocManager::install(shinythemes)
BiocManager::install(shinycssloaders)
devtools::install_github("areyesq89/SummarizedBenchmark", ref = "fdrbenchmark")
BiocManager::install("benchmarkfdrData2019")Note that newer versions of the SummarizedBenchmark package are not compatible, so make sure to use the specific tagged version above. The benchmarkfdrData2019 Bioconductor package contains the results objects used by the App.
shiny::runGitHub("kdkorthauer/benchmarkfdr-shiny")