Note: The software is not maintened since March 31st, 2022.
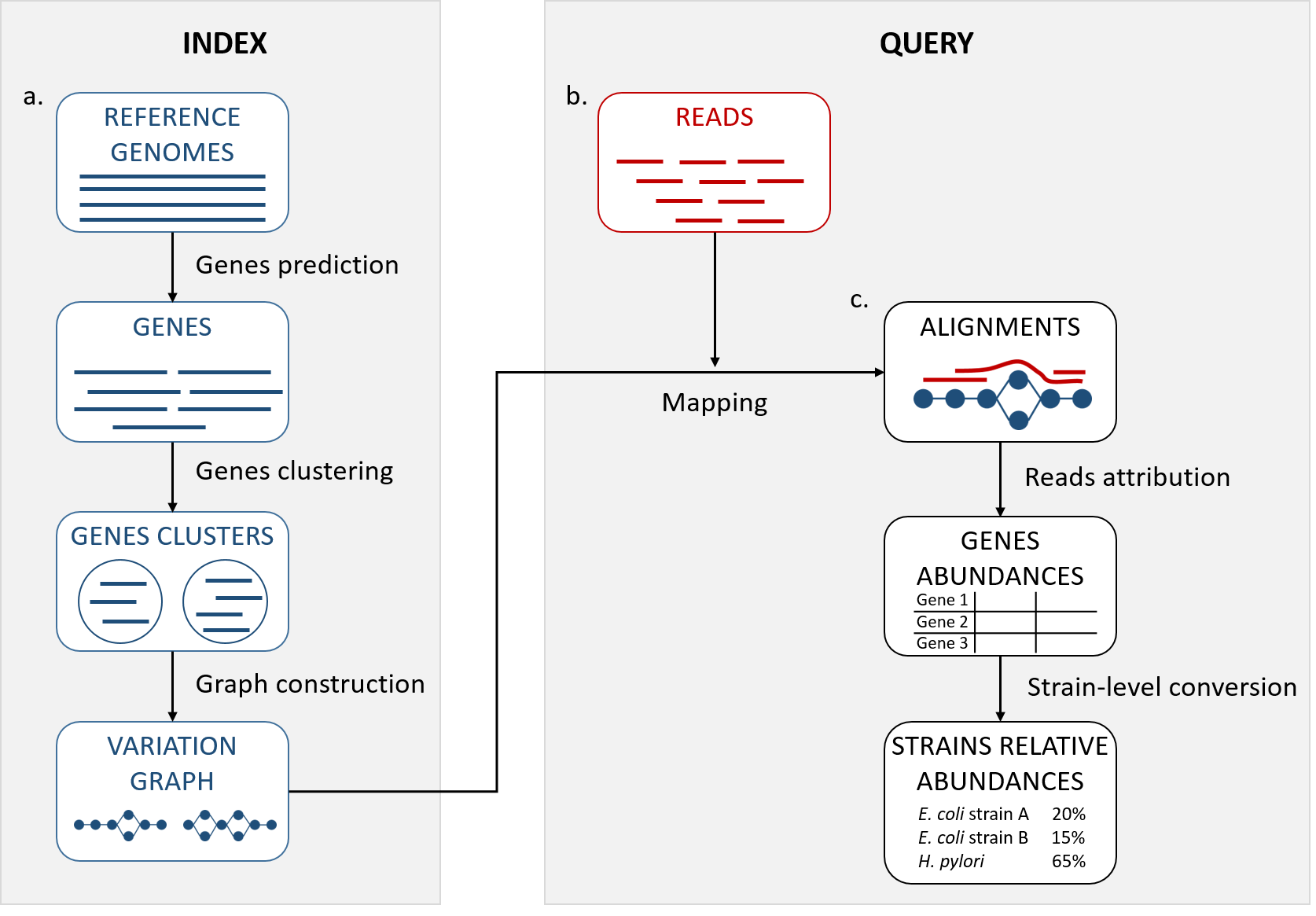
StrainFLAIR (STRAIN-level proFiLing using vArIation gRaph) is a tool for strain identification and quantification that uses variation graph representation of genes sequences. StrainFLAIR is sub-divided into two main parts: first, an indexing step that stores clusters of reference genes into variation graphs, and then, a query step using mapping of metagenomic reads to infere strain-level abundances in the queried sample. The input for indexation is a collection of complete genomes, draft genomes or metagenome-assembled genomes from which genes will be predicted. The input for query is the so-created graph and any read file.
StrainFLAIR is composed of several modules. Each module is described below.
Note StrainFLair dependancies are not satisfied on mac environment. Hence install is currently possible on unix environments only.
Installation may be done with this commands:
git clone https://github.com/kevsilva/StrainFLAIR
cd StrainFLAIR
conda env create -p Strain --file env.yml
conda activate ./Strain
pip install ../StrainFLAIR
git clone --recursive https://github.com/ekg/seqwish.git
cd seqwish
git checkout af8354bdb38a075f813d4cc0ad81cb4f038d1be4
cmake -H. -Bbuild && cmake --build build -- -j 3
cd ..
cp seqwish/bin/seqwish Strain/bin/StrainFLAIR.sh is a pipeline combining the indexation and query steps. Mapping has to be done separately.
Usage: ./StrainFLAIR.sh [index/query]
After the indexation step, predicted genes fasta files are stored in directory_output_name/, clusters files in directory_output_name/clusters/, the graph and its indexes in directory_output_name/graphs/.
Usage: ./StrainFLAIR.sh index -i file_of_files -o directory_output_name [OPTIONS]
MANDATORY
-i <file name of a file of file(s) or of a fasta file>
In case of a fasta file: each fasta input line is considered as a genome
In case of a .txt file: each line contains a fasta file, and each of these fasta is considered as a genome. In this case a genome can span several line, for instance for perfectly assembled genomes
-o <directory_output_name>. This directory must not exist. It is created by the program. All results are stored in this directory
OPTIONS
-l value <int value>. Set the length of the sequences on the left and right part of each predicted gene, added to the indexation graph. [default: 75]
-c value <float value>. Sequence identity threshold [default: 0.95]
-aS value <float value>. Alignment coverage for the shorter sequence [default: 0.90]
-g 0 or 1. [default: 1]
If set to 0, a sequence is clustered to the first cluster that meet the threshold.
If set to 1, a sequence is clustered to the most similar cluster that meet the threshold.
-d value <int value>. Length of description in .clstr file [default: 0]
-M value <int value>. Memory limit (in MB) ; 0 for unlimited. [default: 0]
-T value <int value>. Number of threads ; with 0, all CPUs will be used. [default: 0]
-G 0 or 1. [default: 0]
If set to 0, use local sequence identity.
If set to 1, use global sequence identity.
-h Prints this message and exit
After the query step, mapping files are stored in directory_output_name/mapping/, and gene-level and strain-level abundances are stored in directory_output_name/results/.
Usage: ././StrainFLAIR.sh query -g graph -f1 reads1 -f2 reads2 -t threads -p dict_clusters -d output_directory_name -o output_files_name [OPTIONS]
MANDATORY
-g <file name of a graph (no format)>
-f1 <single-end reads or pair1 of paired-end reads (fastq or fastq.gz)>
-t <number of threads to use for mapping>
-p <pickle file containing the dictionary of clusters and their genes>
-d <output_directory_name>. Name of the directory in which all files are output.
-o <output_files_name>. Specific name for the output files.
OPTIONS
-f2 <pair2 of paired-end reads (fastq or fastq.gz)>
-s value <float value between [0-1]>. Set the threshold on proportion of detected specific genes. [default=0.5]
-h Prints this message and exit
./StrainFLAIR.sh index -i file_of_files.txt -o myproject
./StrainFLAIR.sh query -g myproject/graphs/all_graphs -f1 myproject/myreads.fastq -p myproject/graphs/dict_clusters.pickle -t 24 -d myproject -o test
StrainFLAIR provides two outputs. The first one is a gene-level abundance table, each line is a colored path of the pangenome graph (and hence a gene) containing in columns key characteristics :
- The corresponding gene identifier.
- For each reference genome, the number of copies of the gene. Since each unique version of a gene is represented once in the graph, whereas it can exist in several copies in the genome (duplicate genes), the counts and abundances computed correspond to the sum of those copies. Keeping track of the number of copies is important to normalize the counts.
- The cluster identifier to which the colored path belongs.
- For unique mapped reads: their raw number and their number normalized by the sequence length.
- For unique plus multiple mapped reads: their raw number and their number normalized by the sequence length.
- The mean abundance of the nodes composing the path. Instead of solely counting reads, we make full use of the graph structure and we propose abundances of each node as explained in the manuscript. Hence, for each colored path, the gene abundance is estimated by the mean of the nodes abundance.
- In order to not underestimate the abundance in case of a lack of sequencing depth (which could result in certain nodes to never be traversed by sequencing reads), StrainFLAIR also outputs the mean abundance without the nodes of the path never covered by a read. The same way reads counts are computed using unique mapped reads only or all mapped reads, the mean abundance with and without these none covered nodes are computed in both situations.
- The ratio of covered nodes, defined as the proportion of nodes from the path which abundance is strictly greater than zero.
The final output is a strain-level abundance table containing the estimated abundance of each reference genome according to different computations and the proportion of specific genes detected for each strain.
From the input reference sequences, protein-coding genes are predicted using Prodigal. To reduce mapping bias at the extremities, predicted genes can be extended on both ends (75 bp by default) if the reference sequence it originates from allows it.
Example: genes_prediction -s file_of_fasta_files.txt -o my_output_directory_name -l 75
Genes are clustered using CD-HIT. Genes are then grouped into gene families and the resulting clusters are composed of similar genes according to the user-defined thresholds and parameters.
Example: cd-hit-est -i my_genes_not_extended.fasta -o clusters_files_name -c 0.95 -aS 0.90 -g 1 -d 0 -M 0 -T 0 -G 0
Module graphs_construction and concat_graphs: building a variation graph representing the gene clusters
Each gene cluster (gene family) is converted into a variation graph. All variation graphs are then concatenated into a single one and indexed.
Example:
graphs_construction -s my_genes_extended.fasta -c cluster_file.clstr -o my_output_directory_name
concat_graphs -i my_input_directory_name -s 1000
vg view final_graph.vg > final_graph.gfa
vg prune final_graph.vg | vg index -g final_graph.gcsa -
vg index -x final_graph.xg final_graph.vg
vg snarls final_graph.vg > final_graph.snarls
Mapping of reads onto a variation graph is done using vg mpmap from vg toolkit. The output needs to be into the JSON format.
Example:
vg mpmap -x final_graph.xg -g final_graph.gcsa -s final_graph.snarls -f my_reads.fastq.gz -t 24 -M 10 -m -L 0 > mapping_output.gamp
vg view -j -K mapping_output.gamp > mapping_output.json
Mapping results are processed according to our developed algorithm to attribute abundances to the reference genes.
Example: json2csv -g final_graph.gfa -m mapping_output.json -p dict_clusters.pickle -o output_file_name
Gene-level abundances are converted into strain-level abundances. Strain abundance is set to zero if not metting the threshold of proportion of detected genes.
Example: compute_strains_abundance -i gene_level_table.csv -o output_file_name -t proportion_detected_genes_threshold
This repository presents two manuals to use StrainFLAIR on example datasets.
MINIMAL_EXAMPLE.md is a manual to apply StrainFLAIR on a minimal set of references to index and reads to query, providing quick results.
COOKBOOK.md is a cookbook allowing to reproduce the results presented in the StrainFLAIR paper on the simulated datasets.
Kévin Da Silva: kevin.da-silva@inria.fr
Pierre Peterlongo: pierre.peterlongo@inria.fr