This is the computational pipeline that takes in FACS based, mito-locus multiplexed sequencing data (Figure 1), and outputs SNVs (including indels) for each mitochondrion in each PCR target region [1], succeeding the previous work based on micropipetting [2]. The output is a comma-delimited table containing per-base information including the position, read depth, reference allele, and frequency of variants (substitution/indel) for single mitochondria. The pipeline further processes the data so that low-Phred-score, reverse-strand or low-depth counts can be removed. Deletions are output alongside substitutions while insertions are tabulated in a separate sheet.
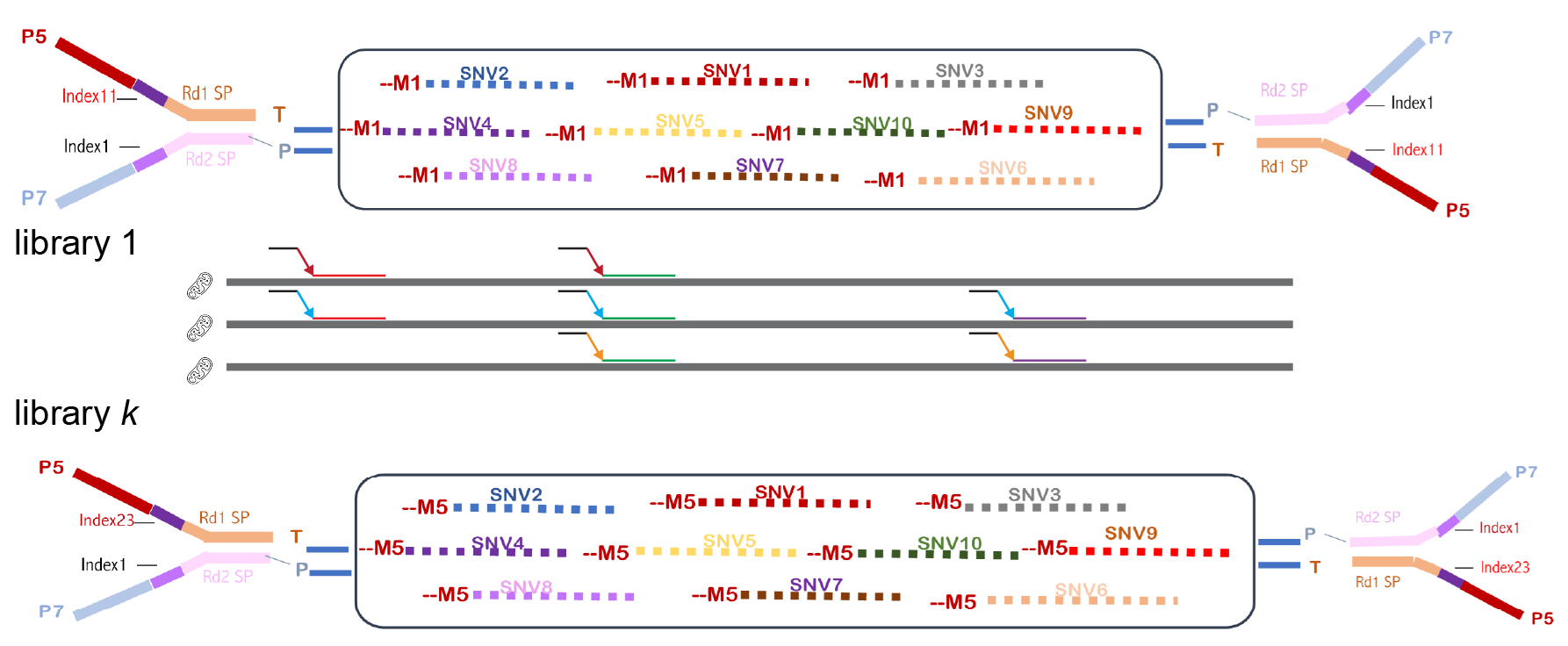 Figure 1. Schematic showing the organelle sorting and multiplexed RCA/PCR amplification technology developed by the Laboratories of David Issadore and James Eberwine (Figure adapted from Hua Zhu; the mitochondrion icon designed by Freepik and obtained from Flaticon).
Figure 1. Schematic showing the organelle sorting and multiplexed RCA/PCR amplification technology developed by the Laboratories of David Issadore and James Eberwine (Figure adapted from Hua Zhu; the mitochondrion icon designed by Freepik and obtained from Flaticon).
- PennSCAP-T preprocessor (>=2.3)
- GNU parallel (>=20190122)
- Perl (>=5.010), String::Approx (>3.0)
- R (>=3.5.0), optparse (>=1.6), parallel (>=4.1)
- Picard (>=2.17)
- samtools (>=1.12)
- Install the dependencies as listed above.
- This package doesn't require installation to system or local
bin. Users just need to download and place the source code folder (Source) into the workplace where the pipeline will be run. Please make sure theDatafolder is the same level asSource. If users want to use some module as standalone, they may simply source the corresponding file in bash or R environment.
- Prepare the sample sheet (
Data/LibraryInfo.csv). The format is exemplified as below
$ head -n 5 Data/LibraryInfo.csv
LibraryID,ExptID,Species,MitoBarcode,SNVLoci,EndType,ReadLength
L1R61P1,777,mouse,M1;M2;M3;M4;M5;M6;M7;M8;M9;M10,v2,SE,150
L2R61P1,777,mouse,M1;M2;M3;M4;M5;M6;M7;M8;M9;M10,v2,SE,150
L3R61P1,777,mouse,M1;M2;M3;M4;M5;M6;M7;M8;M9;M10,v2,SE,150
L4R61P1,777,mouse,M1;M2;M3;M4;M5;M6;M7;M8;M9;M10,v2,SE,150
- Run the pipeline to generate primer and sequencing stats as well as the pileup files by
$ bash Source/01.pipeline.bash
- Generate the SNV table for each sequencing run in R by
source("Source/02.pipeline.R")
Youtao Lu (luyoutao@sas.upenn.edu)
Copyright (c) 2021-2023, Youtao Lu, Erik Nordgren, Stephen Fisher and Junhyong Kim, Department of Biology, University of Pennsylvania
Copyright (c) 2021-2023, Parnika Kadam, Hua Zhu and James Eberwine, Perelman School of Medicine, University of Pennsylvania
Copyright (c) 2021-2023, Zijian Yang, Yasemin Atiyas, Nishal Shah and David Issadore, Penn Engineering, University of Pennsylvania
- Parnika Kadam, Zijian Yang, Youtao Lu, Hua Zhu, Yasemin Atiyas, Nishal Shah, Stephen Fisher, Erik Nordgren, Junhyong Kim, David Issadore and James Eberwine. "Single-Mitochondrion Sequencing Uncovers Distinct Mutational Patterns and Heteroplasmy Landscape in Mouse Astrocytes and Neurons" (BMC Biology 22, 162 (2024))
- Jacqueline Morris, Young-Ji Na, Hua Zhu, Jae-Hee Lee, Hoa Giang, Alexandra V. Ulyanova, Gordon H. Baltuch, Steven Brem, H. Issac Chen, David K. Kung, Timothy H. Lucas, Donald M. O'Rourke, John A. Wolf, M. Sean Grady, Jai-Yoon Sul, Junhyong Kim and James Eberwine. "Pervasive Within-Mitochondrion Single-Nucleotide Variant Heteroplasmy as Revealed by Single-Mitochondrion Sequencing." (Cell Reports 21 (10): 2706-13)