Locally smearing noisy regions in Hi-C contact maps as a prelude to differential analyses
Example: Standard Hi-C matrix ratio without (left) and with (right) serpentine with different numbers of iterations.
| Standard log ratio | Serpentine log ratio |
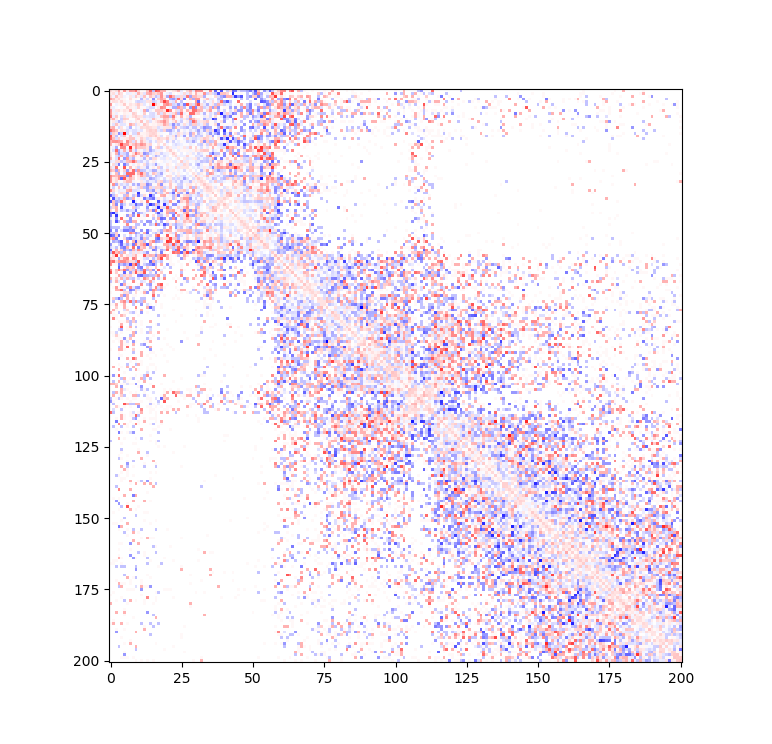 |
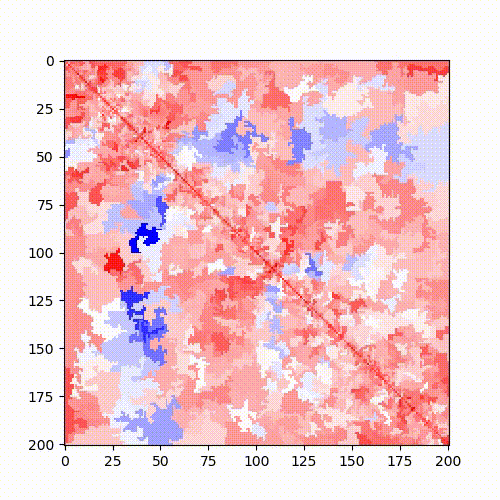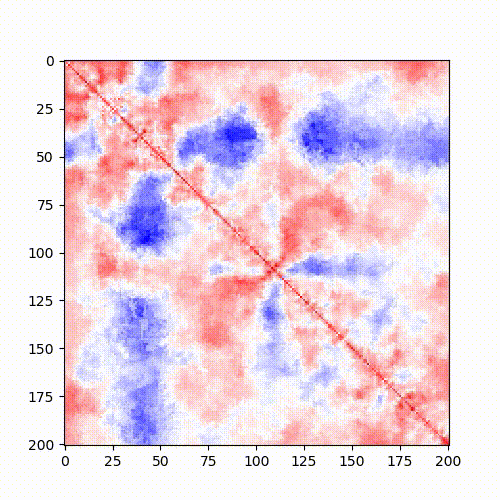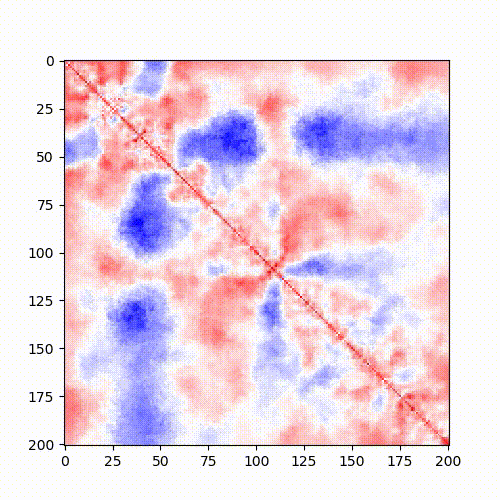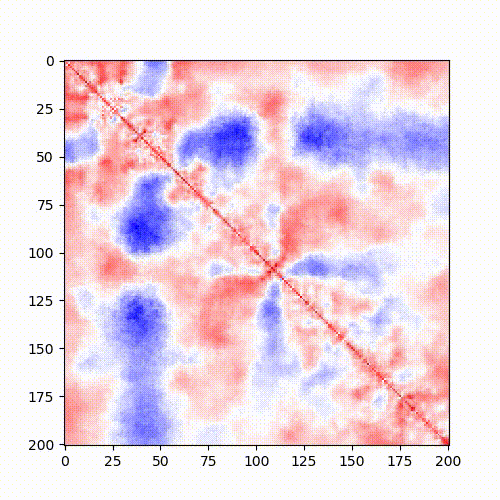 |
Use it as a Python 3 library:
import numpy as np
import serpentine as sp
A = np.loadtxt('./demos/A.csv')
B = np.loadtxt('./demos/B.csv')
trend, threshold = sp.MDbefore(A, B, show=False)
sA, sB, sK = sp.serpentin_binning(A, B, threshold, threshold / 5)Or as a standalone UNIX tool:
$ serpentine --help
Serpentine binning
An implementation of the so-called 'serpentine binning' procedure described
in Baudry et al.
Command line::
Usage:
serpentine.py [<matrixA>] [<matrixB>] [--threshold=auto] [--verbose]
[--min-threshold=auto] [--trend=high] [--triangular]
[--limit=3] [--demo] [--demo-size=500]
Arguments:
matrixA The first input matrix, in plain text
CSV format. Optional in demo mode.
matrixB The second input matrix, in plain text
CSV format. Optional in demo mode or
single binning mode.
Options:
-h, --help Display this help message.
--version Display the program's current version.
-t auto, --threshold auto Threshold value to trigger binning.
[default: auto]
-m auto, --min-threshold auto Minimum value to force trigger binning
in either matrix. [default: auto]
--trend high Trend to subtract to the differential
matrix, possible values are "mean":
equal amount of positive and negative
differences, and "high": normalize
at the regions with higher coverage.
[default: high]
--triangular Treat the matrix as triangular,
useful when plotting matrices adjacent
to the diagonal. [default: False]
--limit 3 Set the z-axis limit on the
plot of the differential matrix.
[default: 3]
--demo Run a demo on randomly generated
matrices. [default: False]
--demo-size 500 Size of the test matrix for the demo.
[default: 500]
-v, --verbose Show verbose output. [default: False]
sudo pip3 install serpentineor, for the very latest version:
sudo pip3 install -e git+https://github.com/koszullab/serpentine.git@master#egg=serpentineExecuting the command serpentine --help will give you a brief help of the command line tool. For a detailed reference to the python library functions, please
read the documentation.
To understand the tool, please read the article on Bioinformatics.
For a full-fledged analysis, consider running the slitherine pipeline.
Vittore F. Scolari (scovit), Lyam Baudry (baudrly), Gael Millot (gmillot)
Baudry L., Millot G. A., Thierry A., Koszul R., and Scolari V.F. Serpentine: a flexible 2D binning method for differential Hi-C analysis. Bioinformatics, 2020 Apr 20, doi: 10.1093/bioinformatics/btaa249
Copyright © 2017-2019 Institut Pasteur, this software has been developed in the Regulation Spatiale des Chromosomes team of Pasteur Institut, Paris, France.
This library is free software; you can redistribute it and/or modify it under the Artistic License.



