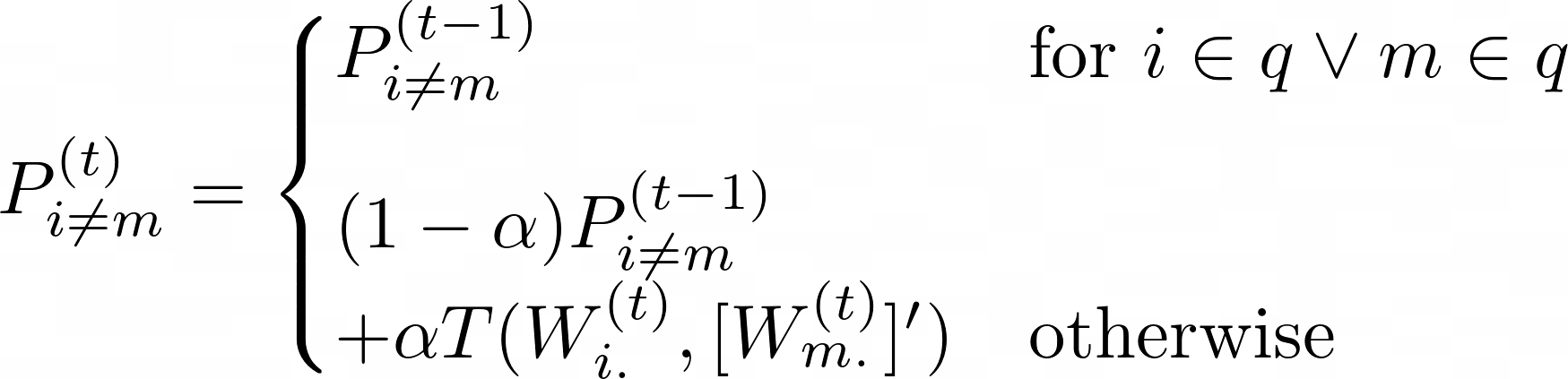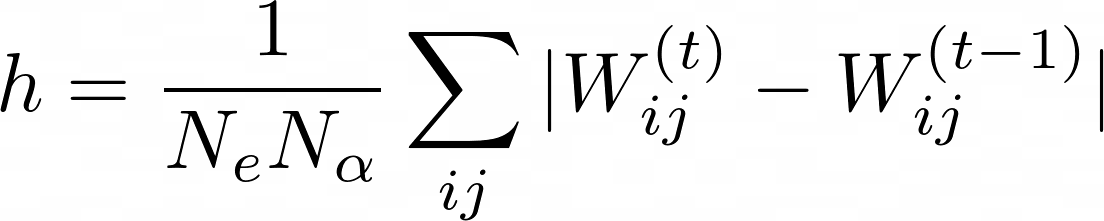Python implementation of the PUMA algorithm.
-
PUMA (PANDA Using MicroRNA Associations)
Manuscript in preparation, used in Hill et al..
C and MATLAB code: https://github.com/mararie/PUMA -
PANDA (Passing Attributes between Networks for Data Assimilation) Glass K, Huttenhower C, Quackenbush J, Yuan GC. Passing Messages Between Biological Networks to Refine Predicted Interactions, PLoS One, 2013 May 31;8(5):e64832
Original PANDA C++ code: http://sourceforge.net/projects/panda-net/. -
LIONESS (Linear Interpolation to Obtain Network Estimates for Single Samples) Kuijjer ML, Tung M, Yuan GC, Quackenbush J, Glass K. Estimating sample-specific regulatory networks, iScience, 2019 Apr 26;14:226-240
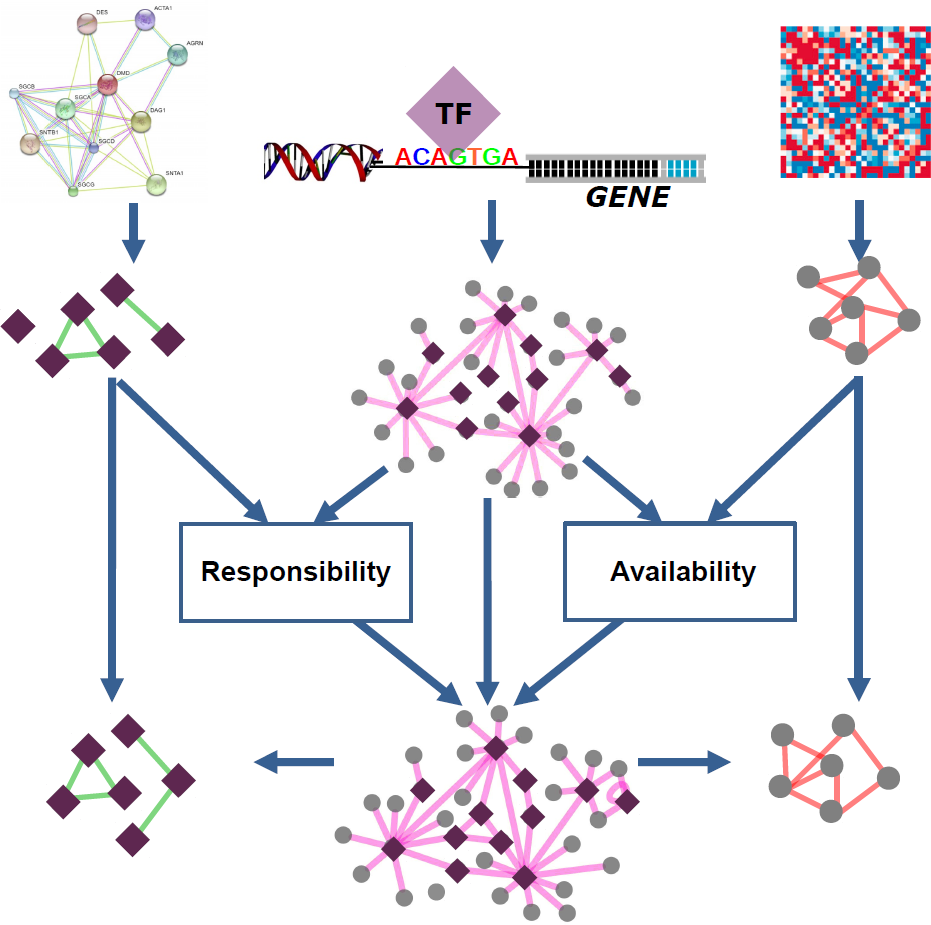
PUMA starts with a prior network of putative regulatory interactions (center network in the image below), a prior network of protein-protein interactions between transcription factors (optional input), and target gene expression data, which is converted into a co-expression network.
A message passing framework is used to find agreement between the three input networks. First, the responsibility (R) is calculated:
as well as the availability (A):
The prior gene regulatory network W is then updated using the responsibility and availability:
Next, the protein cooperativity and gene co-regulatory networks are updated, except for cooperativity interactions made by miRNAs (q):
.Self-interactions in P and C are also updated to satisfy convergence:
, which is evaluated using a hamming distance:
PyPuma runs on Python 3.x. You can either run the PyPuma script directly (see Usage) or install it on your system. We recommend the following commands to install PyPuma on UNIX systems:
Using python virtual environments is the cleanest installation method.
Cloning git and setting up a python virtual environment:
pip install --user pipenv #Make sure you have pipenv
git clone https://github.com/aless80/PyPuma.git
cd PyPuma
Creating a virtual environment and installing PyPuma:
virtualenv pypumaenv #virtual environment created in a folder inside the git folder
source pypumaenv/bin/activate
(pypumaenv)$ pip install -r requirements.txt
(pypumaenv)$ python setup.py install --record files.txt
Uninstall PyPuma from virtual environment:
cat files.txt | xargs rm -rf
Complete removal of virtual environment and PyPuma:
(pypuma)$ deactivate #Quit virtual environment
rm -rf pypumaenv
Never use sudo pip. Instead you can use pip on the user's install directory:
git clone https://github.com/aless80/PyPuma.git
cd PyPuma
python setup.py install --user
#to run from the command line you will need to make PyPuma executable and add the bin directory to your PATH:
cd bin
chmod +x PyPuma
echo "$(pwd):PATH" >> ~/.bashrc
source ~/.bashrc
To run PyPuma from Windows (not fully tested) install Git (https://git-scm.com/downloads) and Anaconda (https://www.continuum.io/downloads) and from the Anaconda prompt run:
git clone https://github.com/aless80/PyPuma.git
cd PyPuma
python setup.py install
PyPuma can be run directly from the terminal with the following options:
-h help
-e, --expression: expression values
-m, --motif: pair file of motif edges, or Pearson correlation matrix when not provided
-p, --ppi: pair file of PPI edges
-o, --output: output file
-i, --mir: mir data miR file
-r, --rm_missing
-q, --lioness: output for Lioness single sample networks
To run PyPuma on the included Toy example:
python run_puma.py -e ./ToyData/ToyExpressionData.txt -m ./ToyData/ToyMotifData.txt -p ./ToyData/ToyPPIData.txt -i ToyData/ToyMiRList.txt -o output_puma.txt
To run LIONESS on PUMA networks, use the flag -q (note that this can take a long time and use considerable computing resources):
python run_puma.py -e ./ToyData/ToyExpressionData.txt -m ./ToyData/ToyMotifData.txt -p ./ToyData/ToyPPIData.txt -i ToyData/ToyMiRList.txt -o output_puma.txt -q output_lioness.txtFinally, note that running PUMA without importing motif and expression data will estimate a co-expression network using Pearson correlation.
Fire up your python shell or ipython notebook. Import the classes in the PyPuma library:
from pypuma.puma import Puma
from pypuma.lioness import LionessThen run PUMA:
puma_obj = Puma('ToyData/ToyExpressionData.txt', 'ToyData/ToyMotifData.txt', 'ToyData/ToyPPIData.txt','ToyData/ToyMiRList.txt')Save the results:
puma_obj.save_puma_results('Toy_Puma.pairs.txt')Example of returning a network visualization of the top edges:
puma_obj.top_network_plot(top=70, file='top_genes.png')Calculate indegrees for further analysis:
indegree = puma_obj.return_puma_indegree()Calculate outdegrees for further analysis:
outdegree = puma_obj.return_puma_outdegree()Run the LIONESS algorithm for single sample networks:
lioness_obj = Lioness(puma_obj)Save LIONESS results:
lioness_obj.save_lioness_results('Toy_Lioness.txt')Return a network plot for one of the LIONESS single sample networks:
plot = AnalyzeLioness(lioness_obj)
plot.top_network_plot(column= 0, top=100, file='top_100_genes.png')The example gene expression data that we have available here contains gene expression profiles for different samples in the columns. Of note, this is just a small subset of a larger gene expression dataset. We provided these "toy" data so that the user can test the method.
However, if you plan to model gene regulatory networks on your own dataset, you should use your own expression data as input.