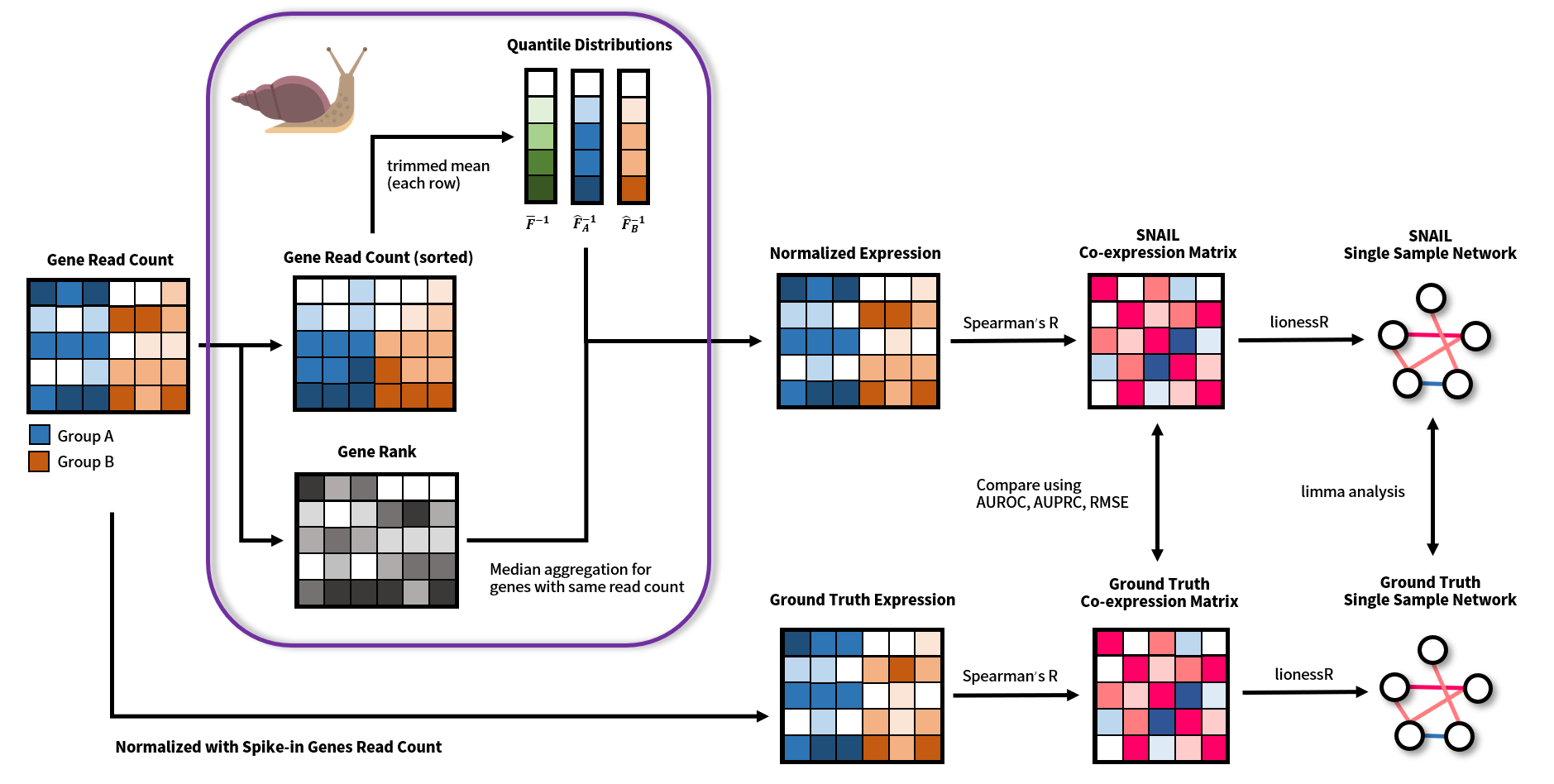
Smooth-quantile Normalization Adaptation for Inference of co-expression Links (SNAIL) is a method that corrects for false-positive associations, which may form between lowly expressed genes after smooth quantile normalization of the data, and which may affect downstream co-expression network analysis. The PySNAIL software is developed by the Kuijjer Lab.
SNAIL is an alternative implementation of smooth quantile normalization (qsmooth), designed to correct false-positive gene associations in qsmooth-normalized RNA-Seq data. Specifically, instead of using the mean value of counts to normalize the expression of genes with the same read count, as is done in the original qsmooth algorithm, SNAIL uses median aggregation (step 7 and step 8 in the following figure). We developed PySNAIL as a standalone Python package supporting multi-thread optimization. We've also implemented a diagnostic function that computes the proportion of affected genes for each sample, to help detect whether smooth quantile normalization would introduce false-positive co-expression between genes in a specific dataset.
It is highly recommended to use the conda virtual environment to install this package. After installation of conda, create a virtual environment for PySNAIL using the following command:
$ conda create -n pysnail python=3.7.7
$ conda activate pysnailDownload the source code from git@github.com:kuijjerlab/PySNAIL.git and install the package using pip:
$ git clone git@github.com:kuijjerlab/PySNAIL.git
$ cd PySNAIL
$ pip install -e .To reproduce the analysis we provide in the manuscript (we use pip to install the Python dependency simply to improve the efficiency of solving conflicts between packages):
$ git clone git@github.com:kuijjerlab/PySNAIL.git
$ cd PySNAIL
$ conda activate pysnail
$ conda config --add channels anaconda
$ conda config --add channels defaults
$ conda config --add channels bioconda
$ conda config --add channels conda-forge
$ conda config --add channels bokeh
$ conda install --file conda_requirement.txt
$ pip install -r pip_requirement.txt
$ pip install -e .The PySNAIL packages includes an example dataset for users to test the method on. This can be found under the test directory. The expression.tsv contains expression levels for 10,000 randomly selected genes from the Mouse ENCODE Project Consortium. The groups.tsv contains the tissue information for each sample.
$ sample_data/expression.tsv
$ sample_data/groups.tsvAfter installation, PySNAIL can be executed directly as a Python module using the following command:
$ pysnail sample_data/expression.tsv --groups sample_data/groups.tsv --outdir output
The complete arguments are listed as follows (one can get this information by executing pysnail --help)
pysnail -h
usage: pysnail [-h] [-g [path]] [-m {'mean', 'median', 'auto'}]
[-t [threshold]] [-o [path]] xprs
Python implementation of Smooth-quantile Normalization Adaptation for
Inference of co-expression Links (PySNAIL)
positional arguments:
xprs Path to the expression data. The file should be
formatted as follows: the rows should represent genes
(the first row must be the sample names), and the
columns should represent samples (the first column
must be the gene names). The columns must be separated
with <tab>.
optional arguments:
-h, --help show this help message and exit
-g [path], --groups [path]
Path to the group information for each sample. The
file should have two columns without any header. The
first column should be the sample names, corresponds
to the columns in xprs. The second column should be
the group information for each sample. The two columns
must be separated by <tab>. If this argument is not
provided, the algorithm will treat all samples as one
group.
-m {'mean', 'median', 'auto'}, --method {'mean', 'median', 'auto'}
Method used compute the aggregate statistics for
quantile with same value in each group, should be
either 'mean', 'median' or 'auto'. If set to 'auto',
the algorithm is going to use median aggregation if
the proportion of the affected samples is larger or
equal to [--threshold] (default: 0.25). Default:
'median'.
-t [threshold], --threshold [threshold]
Threshold of the proportion of samples being affected
if mean aggregation is being used. The algorithm is
going to use median aggregation if the proportion of
the affected samples is larger or equal to this
threshold when [--method] is set to 'auto'. This
argument is ignored if method is specified with 'mean'
or 'median'. Default: 0.25
-c [cutoff], --cutoff [cutoff]
Cutoff used for trimmed mean when inferring quantile
distribution. (range from 0.00 to 0.25) Default: 0.15.
-o [path], --outdir [path]
Output directory for the corrected qsmooth expression
and some informative statistics. The directory
consists of a data table 'xprs_norm.tsv' with the
corrected expression levels. Default: './output'.
The bioconductor-encodexplorer package used in the original analysis is deprecated. To reproduce the analysis, please download the ENCODE dataset from here before executing the following commands.
To reproduce analysis in the manuscript:
$ cd PySNAIL
$ # download the ZIP file and put it here.
$ mkdir -p manuscript_analysis/datasets/
$ unzip PySNAIL-ENCODE.zip
$ mv ENCODE manuscript_analysis/datasets/
$ snakemake --cores [n]The result can be found in the directory manuscript_analysis. Note that it will likely take a while to download and preprocess the datasets.
PySNAIL also provides an application programming interface (API) in Python for developers or bioinformaticians who wants to control more parameters used in the analysis.
import os
from pysnail import Dataset, qsmooth
xprs = os.path.realpath('sample_data/expression.tsv')
groups = os.path.realpath('sample_data/groups.tsv')
dataset = Dataset(xprs, groups, **{'index_col': 0, 'sep': '\t'})
xprs_norm, qstat = qsmooth(dataset, aggregation='auto', threshold=0.2)
xprs_norm.to_csv(os.path.realpath('pysnail_out.tsv'), sep='\t')print(analysis.dataset)Get number of affected genes for each samples.
qstat.num_affected_genesResult:
Group Sample
Embryonic Facial Prominence ENCFF132NQU 0
ENCFF262TXH 0
ENCFF369TLJ 0
ENCFF370UDF 0
ENCFF536XKZ 0
...
Stomach ENCFF052DOQ 0
ENCFF288JNN 0
ENCFF355MOU 0
ENCFF691EQW 0
ENCFF972NMO 2730
Length: 126, dtype: int64Get number of affected samples in the dataset.
qstat.num_affected_samples # 1Get affected genes for each sample.
print(qstat.num_affected_samples)Result:
Group Embryonic Facial Prominence \
Sample ENCFF132NQU ENCFF262TXH ENCFF369TLJ
ENSMUSG00000082905 False False False
ENSMUSG00000026174 False False False
ENSMUSG00000031293 False False False
ENSMUSG00000062458 False False False
ENSMUSG00000083793 False False False
... ... ... ...
ENSMUSG00000015093 False False False
ENSMUSG00000098607 False False False
ENSMUSG00000102632 False False False
ENSMUSG00000093969 False False False
ENSMUSG00000050876 False False False
Group \
Sample ENCFF370UDF ENCFF536XKZ ENCFF594CEM ENCFF672DDJ
ENSMUSG00000082905 False False False False
ENSMUSG00000026174 False False False False
ENSMUSG00000031293 False False False False
ENSMUSG00000062458 False False False False
ENSMUSG00000083793 False False False False
... ... ... ... ...
ENSMUSG00000015093 False False False False
ENSMUSG00000098607 False False False False
ENSMUSG00000102632 False False False False
ENSMUSG00000102632 False False False False
ENSMUSG00000093969 False False False False
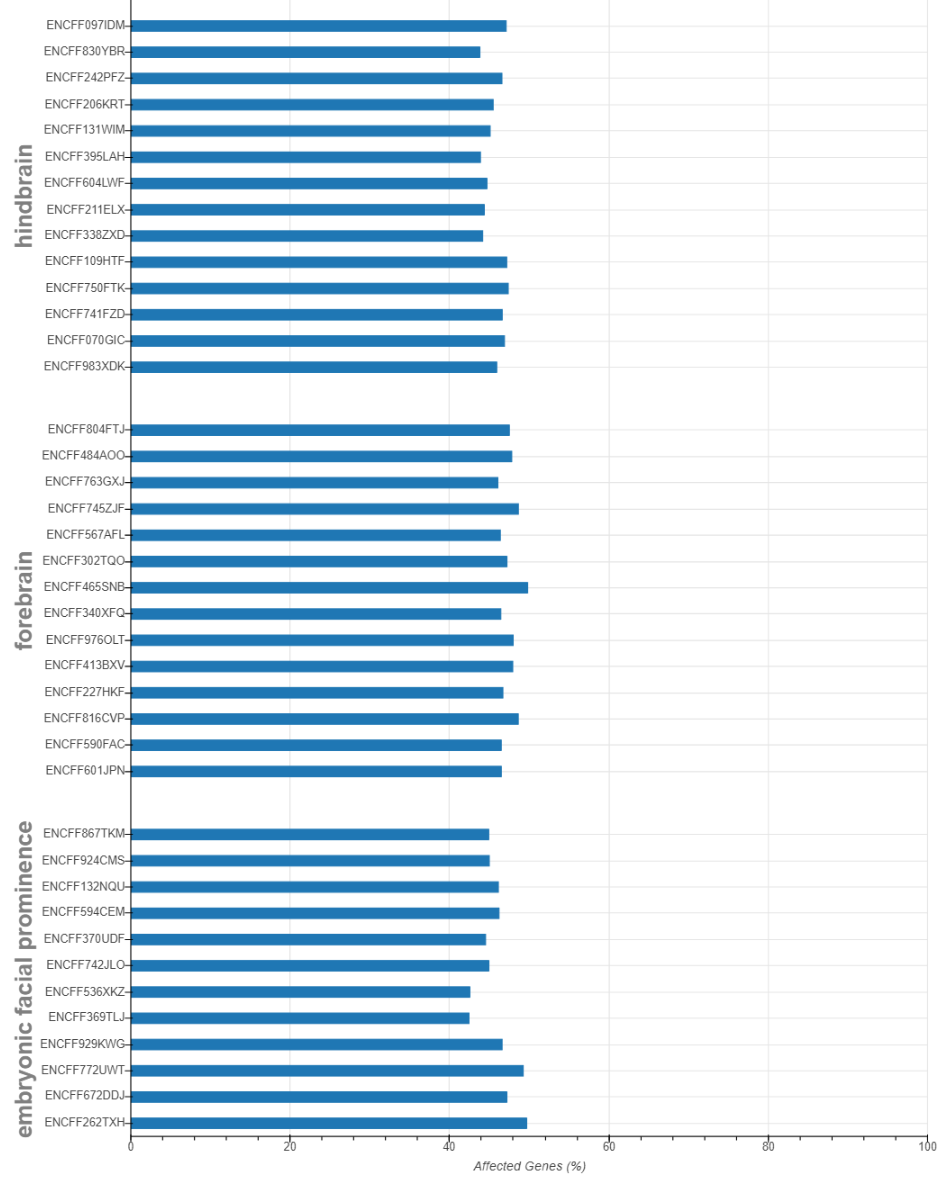
ENSMUSG00000050876 False False False False Make bar plot on number of affected genes for each sample
from pysnail import bokeh_affected_barplot
bokeh_affected_barplot(dataset, qstat, 'output')* The result shown here is excerpted from the analysis in the manuscript instead of from the sample data.
For a more detailed description, please refer the the documentation of PySNAIL.