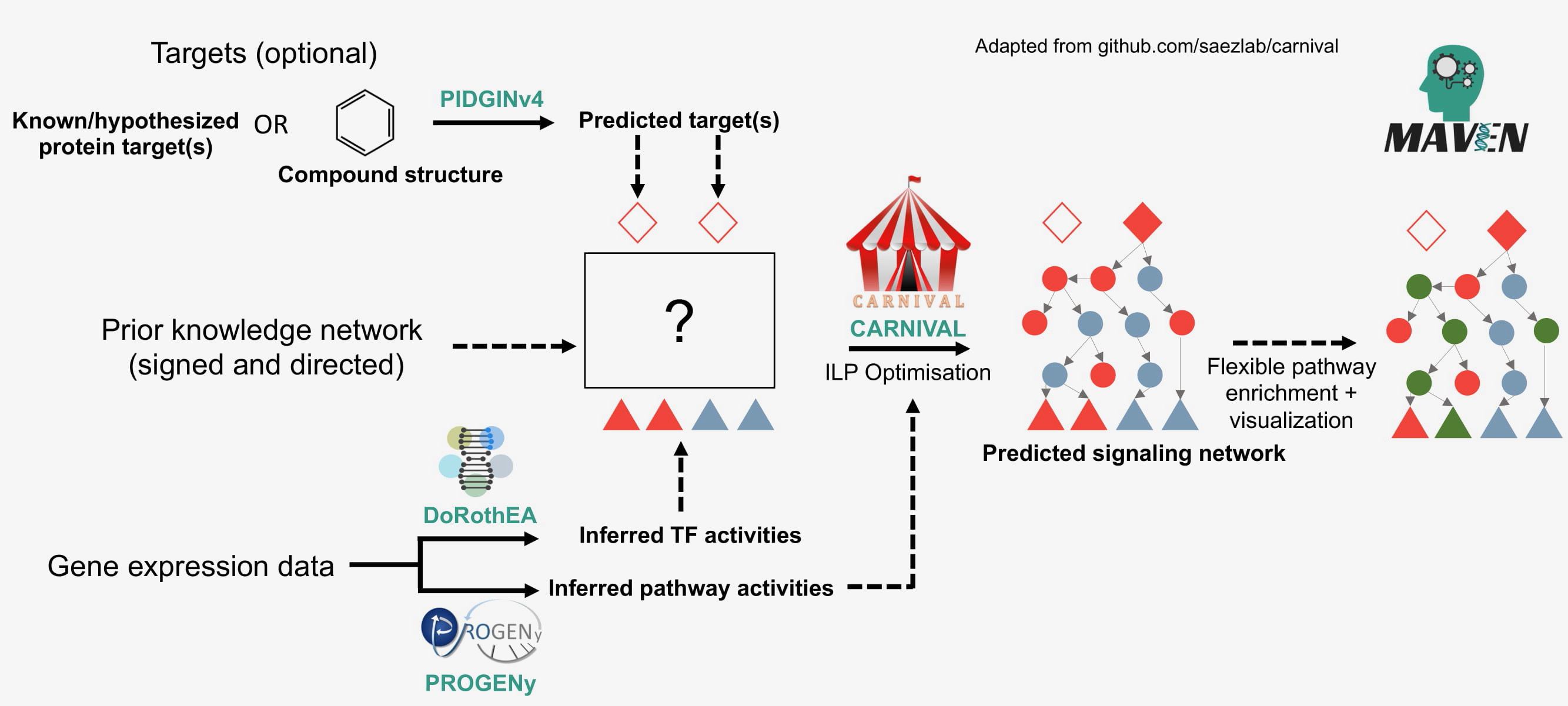
MAVEN is an R shiny app which enables integrated bioinformatics and chemoinformatics analysis for mechansism of action analysis and visualisation.
The tool is a collaborative work between the Bender Group at University of Cambridge and Saez Lab at Heidelberg University (Rosa Hernansaiz Ballesteros https://github.com/rosherbal).
Documentation can be found at https://laylagerami.github.io/MAVEN/. MAVEN can be installed locally or as a Docker or Singularity container. For a step-by-step tutorial for exploring the mechanism of action of a query compound, you can view our example using the HER2 inhibitor lapatanib.
Implemented approaches and data:
- Target prediction provided by PIDGIN (BenderGroup/PIDGINv4)
- Prior knowledge network from Omnipath DB (https://omnipathdb.org/)
- SMILES widgets provided by ChemDoodle (zachcp/chemdoodle)
- TF enrichment with DoRothEA (saezlab/dorothea)
- Pathway analysis with PROGENy (saezlab/progeny)
- Causal reasoning with CARNIVAL (saezlab/carnival)
- MSigDb gene sets for network pathway enrichment (http://www.gsea-msigdb.org/gsea/msigdb/index.jsp)
- Helper scripts are provided by saezlab/transcriptutorial
Check out https://github.com/saezlab/shinyfunki for a multi-omic functional integration and analysis platform which implements many of the same tools.
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see https://www.gnu.org/licenses/.