The main goal of genefam_dist is to describe the "tree signal" of a gene family, which is a vector of 'scores' describing how well supported this gene family is by a set of reference species trees. You can download here (PDF) a low-resolution version of the poster presented at Evolution 2018.
The software is composed of a (low-level) library to handle mul-trees, and both a python module and standalone programs using this library.
The python module is called treesignal and is the standard way of creating the tree signatures. It uses
dendropy for tree manipulation and scikit-learn for storing the signature data into
a tidy data set. The standalone programs are described below.
The library is written in C, with functions to calculate distances between mul-trees (gene families) and species trees. It is a branched version of the internal biomcmc library which is part of the guenomu software for phylogenomic species tree inference.
The software should be installed in two stages: first the genefam_dist library is compiled and installed (only once, maybe globally). Then the python
module can be installed (one per python version, for instance). I provide below one example of installation (change as appropriate).
The C library (and the standalone programs) use the autotools build system. Assuming you downloaded the zip file for the master version:
/home/simpson/$ unzip genefam-dist-master.zip
/home/simpson/$ mkdir build
/home/simpson/$ cd build
/home/simpson/$ ../genefam_dist-master/configure --prefix /usr/local
/home/simpson/$ make; sudo make install
As seen above, it is usually good idea to compile the code on a dedicated clean directory (build, in the example).
The example above will install the libgenefamdist family globally, in /usr/local. If you don't have administrative priviledges you can
chose a local directory (and drop the "sudo" command).
The python module can be installed with
/home/simpson/$ cd ../genefam_dist-master/python
/home/simpson/genefam_dist-master/python$ vi setup.py ### here you have to add the prefix by hand into the parameters
/home/simpson/genefam_dist-master/python$ python3 setup.py build;
/home/simpson/genefam_dist-master/python$ python3 setup.py install --user
The parameters that need to be changed are include_dirs, library_dirs, and runtime_library_dirs, and they need to point to the
include and lib sudirectories of the library installation (e.g. include_dirs = /usr/local/include).
Yes, this is less than ideal, I will fix that (probably by making autotools generate this file...)
After that, you can use the module in your python code by calling import treesignal --- see notebook (ipynb) examples in the docs directory.
Here is a list of standalone programs that will be compiled and installed with autotools. Run them without arguments for a brief explanation (they are simple but require a rigid structure for the arguments).
-
gf_distmatrix_genetree_sptree given two files, one with a list of gene trees and one with a list of species trees, calculates a set of distances between all tree pairs
-
gf_distsignal_genetree_sptree given two files, like the distmatrix program above, but all distances for one gene tree per line, as in the tree signature vector
-
gf_generate_spr_trees generates a series of random trees with a given number of SPR operations apart from the next one.
-
gf_spr_distance calculates the approximate unrooted SPR distance between consecutive pairs of trees in a nexus file (this program assumes single-labelled trees, that is, no paralogs etc. are allowed)
-
gf_concatenate_trees takes two nexus trees (from e.g. MrBayes analyses) and merge them by excluding first trees and thinning, if needed. It also excludes less frequent trees and assumes unrooted topologies (i.e. it neglects root information), important since returns a trprobs nexus file.
-
gf_find_best_trees takes a (large) number of gene families and estimates patristic distance-based species trees, as well as tries to find species trees minimising all/some distances. This is an experimental program, intended as a replacement for the
bmc2_maxtreeprogram from guenomu.
More details to follow,
- For running the programs, you can have a brief description by running them with no parameters.
- For more details about the calculations and theory, please check the guenomu software and the accompanying publication
- A Bayesian Supertree Model for Genome-Wide Species Tree Reconstruction, Systematic Biology, 2016 DOI: 10.1093/sysbio/syu082
- If you have any question, comment or request please do not hesitate to contact me at leomrtns@gmail.com
- For an (not extensive) introduction on tree distances that I will be focusing on, please have a look at my recent talk at UNIL.
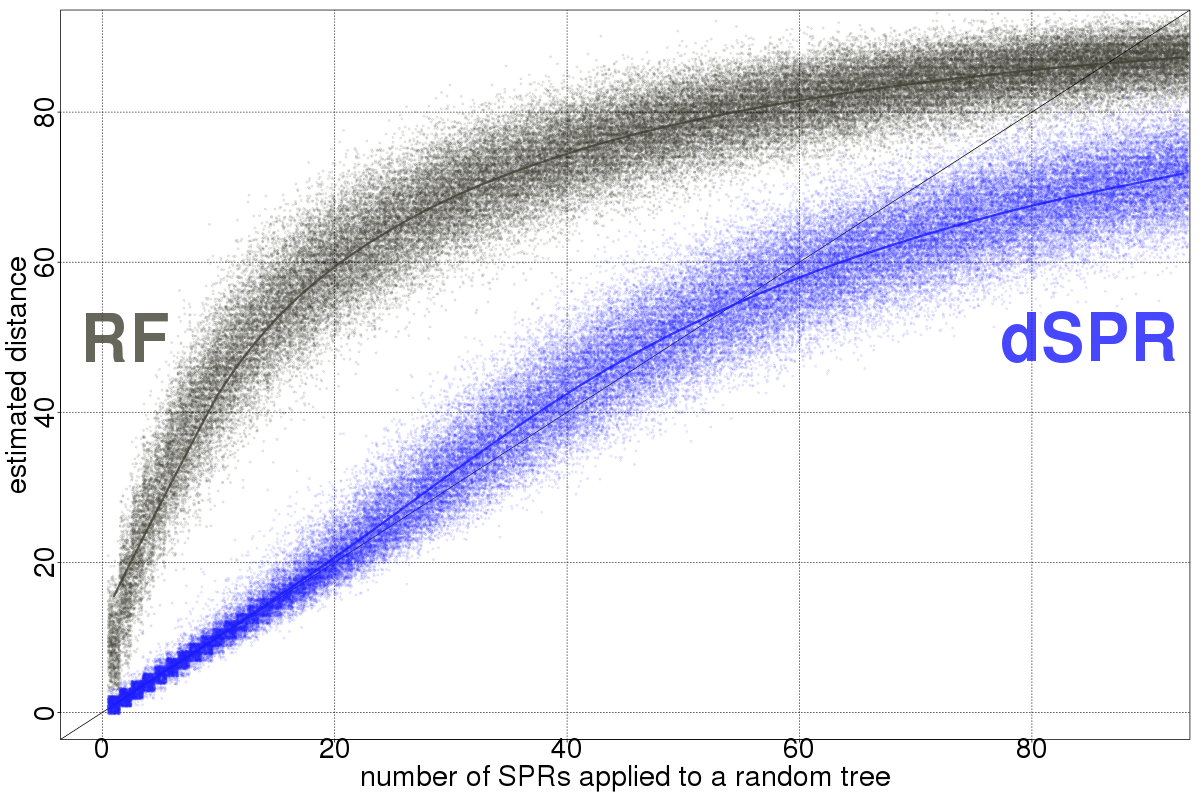
The dSPR is an approximation to the unrooted SPR which was used on the recombination detection model
(but faster than the original version and capable of working with arbitrarily large trees).
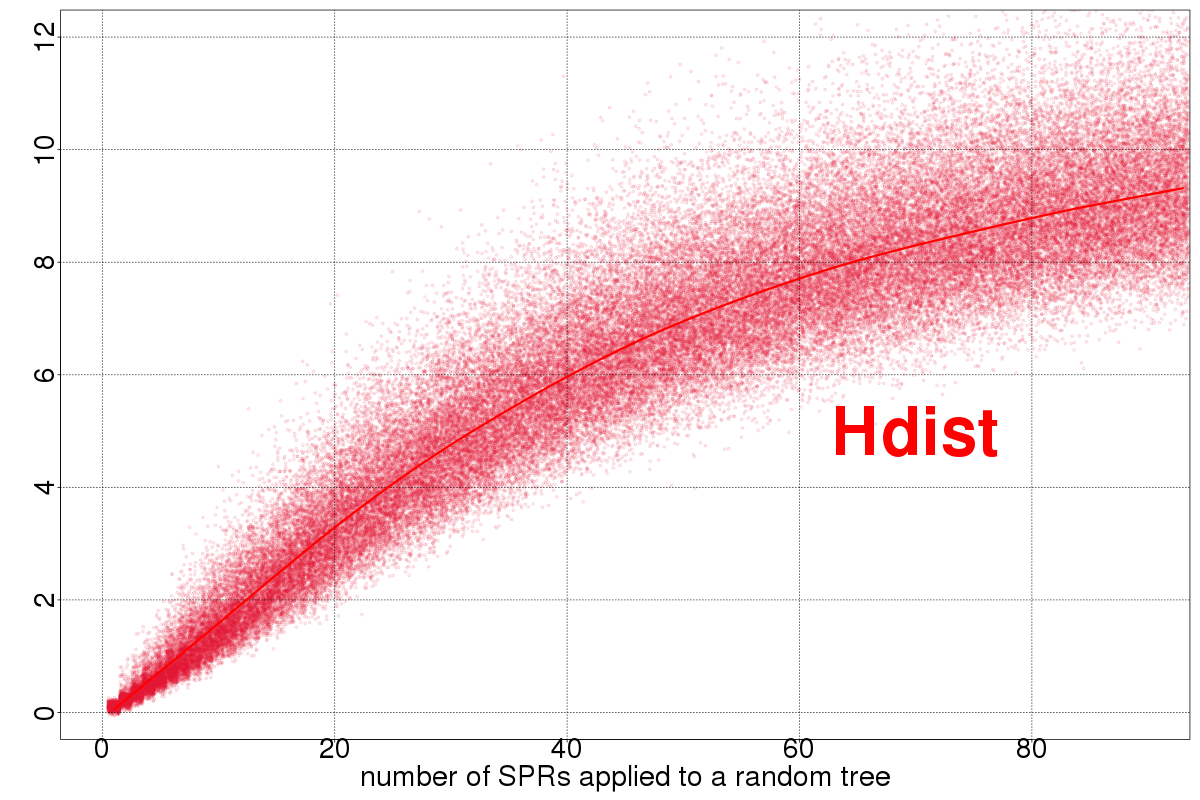
The Hdist is related to the clade alignment score, that I rediscovered (with modifications) when speeding up the
dSPR algortihm.
Notice that our current implementations of these distances can work on mul-trees on a very experimental setting (using the extended species tree concept from the mulRF algorithm), but the results below assume the same leaf set between trees.
Copyright (C) 2016-today Leonardo de Oliveira Martins
genefam-dist is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version (http://www.gnu.org/copyleft/gpl.html).