This package contains various methods to normalize the intensity of various modalities of magnetic resonance (MR) images, e.g., T1-weighted (T1-w), T2-weighted (T2-w), FLuid-Attenuated Inversion Recovery (FLAIR), and Proton Density-weighted (PD-w).
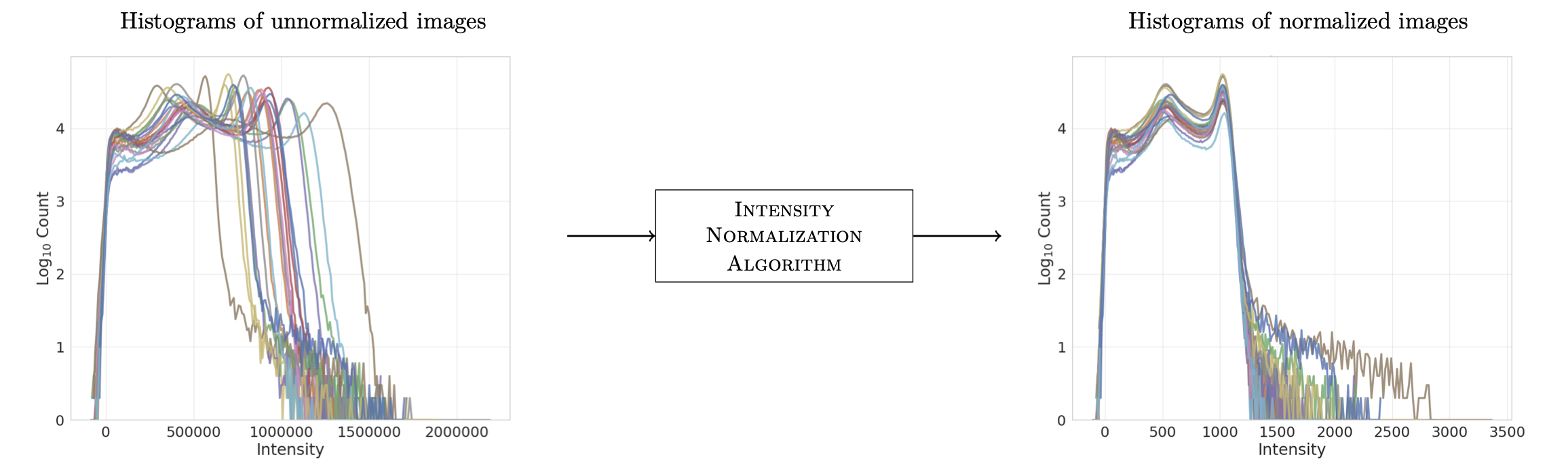
The basic functionality of this package can be summarized in the following image:
where the left-hand side are the histograms of the intensities for a set of unnormalized images (from the same scanner with the same protocol!) and the right-hand side are the histograms after (FCM) normalization.
We used this package to explore the impact of intensity normalization on a synthesis task (pre-print available here).
Note that while this release was carefully inspected, there may be bugs. Please submit an issue if you encounter a problem.
This package was developed by Jacob Reinhold and the other students and researchers of the Image Analysis and Communication Lab (IACL).
We implement the following normalization methods (the names of the corresponding command-line interfaces are to the right in parentheses):
- Z-score normalization (
zscore-normalize) - Fuzzy C-means (FCM)-based tissue-based mean normalization (
fcm-normalize) - Gaussian Mixture Model (GMM)-based WM mean normalization (
gmm-normalize) - Kernel Density Estimate (KDE) WM mode normalization (
kde-normalize) - WhiteStripe [3] (
ws-normalize)
- Least squares (LSQ) tissue mean normalization (
lsq-normalize) - Piecewise Linear Histogram Matching (Nyúl & Udupa) [1,2] (
nyul-normalize) - RAVEL [4] (
ravel-normalize)
Individual image-based methods normalize images based on one time-point of one subject.
Sample-based methods normalize images based on a set of images of (usually) multiple subjects of the same modality.
Recommendation on where to start: If you are unsure which one to choose for your application, try FCM-based WM-based normalization (assuming you have access to a T1-w image for all the time-points). If you are getting odd results in non-WM tissues, try least squares tissue normalization (which minimizes the least squares distance between CSF, GM, and WM tissue means within a set).
All algorithms except Z-score (zscore-normalize) and the Piecewise Linear Histogram Matching (nyul-normalize) are
specific to images of the brain.
Intensity normalization is an important pre-processing step in many image processing applications regarding MR images since MR images have an inconsistent intensity scale across (and within) sites and scanners due to, e.g.,:
- the use of different equipment,
- different pulse sequences and scan parameters,
- and a different environment in which the machine is located.
Importantly, the inconsistency in intensities isn't a feature of the data (unless you want to classify the scanner/site from which an image came)—it's an artifact of the acquisition process. The inconsistency causes a problem with machine learning-based image processing methods, which usually assume the data was gathered iid from some distribution.
- matplotlib
- numpy
- nibabel
- scikit-fuzzy
- scikit-learn
- scipy
- statsmodels
We have provided a script create_env.sh to create a conda environment with the necessary packages
(run like: . ./create_env.sh, this package will be installed in the created environment)
The easiest way to install the package is through the following command:
pip install intensity-normalization
OR, via the conda package manager:
conda install -c conda-forge intensity-normalization
To install from the source directory, use
python setup.py install
Note the package antspy is required for the RAVEL normalization routine, the
preprocessing tool as well as the co-registration tool, but all other normalization and processing tools work without
it. To also install the antspy package, run pip install antspyx before installing this package or use
the create_env.sh script with the option --antspy. If the pip installation fails, you may need to build antspy from
source.
You can use the several provided command line scripts to interface with the package, e.g.,
fcm-normalize -i t1/ -m masks/ -o test_fcm -v
where t1/ is a directory full of N T1-w images and masks/ is a directory full of N corresponding brain masks,
test_fcm is the output directory for the normalized images, and -v controls the verbosity of the output.
The command line interface is standard across all normalization routines (i.e., you should be able to run all normalization routines with the same call as in the above example); however, each has unique options and not all methods support single image processing.
Call any executable script with the -h flag to see more detailed instructions about the proper call.
Note that brain masks (or already skull-stripped images) are required for most of the normalization methods. The brain masks do not need to be perfect, but each mask needs to remove most of the tissue outside the brain. Assuming you have T1-w images for each subject, an easy and robust method for skull-stripping is ROBEX [5].
You can install ROBEX—and get python bindings for it at the same time–with the
package pyrobex (installable via pip install pyrobex).
In addition to the above small tutorial, there is consolidated documentation here.
-
This package was developed to process adult human MR images; neonatal, pediatric, and animal MR images should also work but—if the data has different proportions of tissues or differences in relative intensity among tissue types compared with adults—the normalization may fail. The
nyul-normalizemethod, in particular, will fail hard if you train it on adult data and test it on non-adult data (or vice versa). Please open an issue if you encounter a problem with the package when normalizing non-adult human data. -
When we refer to any specific modality, it is referring to a non-contrast version unless otherwise stated. Using a contrast image as input to a method that assumes non-contrast will produce suboptimal results. One potential way to normalize contrast images with this package is to 1) find a tissue that is not affected by the contrast (e.g., grey matter) and normalize based on some summary statistic of that (where the tissue mask was found on a non-contrast image); 2) use a simplistic (but non-robust) method like Z-score normalization.
Unit tests can be run from the main directory as follows:
nosetests -v --with-coverage --cover-tests --cover-package=intensity_normalization tests
If you are using docker, then the equivalent command will be (depending on how the image was built):
docker run jcreinhold/intensity-normalization /bin/bash -c "pip install nose && nosetests -v tests/"
You can build a singularity image from the docker image hosted on dockerhub via the following command:
singularity pull --name intensity_normalization.simg docker://jcreinhold/intensity-normalization
If you use the intensity-normalization package in an academic paper, please cite the
corresponding paper:
@inproceedings{reinhold2019evaluating,
title={Evaluating the impact of intensity normalization on {MR} image synthesis},
author={Reinhold, Jacob C and Dewey, Blake E and Carass, Aaron and Prince, Jerry L},
booktitle={Medical Imaging 2019: Image Processing},
volume={10949},
pages={109493H},
year={2019},
organization={International Society for Optics and Photonics}}
[1] N. Laszlo G and J. K. Udupa, “On Standardizing the MR Image Intensity Scale,” Magn. Reson. Med., vol. 42, pp. 1072–1081, 1999.
[2] M. Shah, Y. Xiao, N. Subbanna, S. Francis, D. L. Arnold, D. L. Collins, and T. Arbel, “Evaluating intensity normalization on MRIs of human brain with multiple sclerosis,” Med. Image Anal., vol. 15, no. 2, pp. 267–282, 2011.
[3] R. T. Shinohara, E. M. Sweeney, J. Goldsmith, N. Shiee, F. J. Mateen, P. A. Calabresi, S. Jarso, D. L. Pham, D. S. Reich, and C. M. Crainiceanu, “Statistical normalization techniques for magnetic resonance imaging,” NeuroImage Clin., vol. 6, pp. 9–19, 2014.
[4] J. P. Fortin, E. M. Sweeney, J. Muschelli, C. M. Crainiceanu, and R. T. Shinohara, “Removing inter-subject technical variability in magnetic resonance imaging studies,” NeuroImage, vol. 132, pp. 198–212, 2016.
[5] Iglesias, Juan Eugenio, Cheng-Yi Liu, Paul M. Thompson, and Zhuowen Tu. "Robust brain extraction across datasets and comparison with publicly available methods." IEEE transactions on medical imaging 30, no. 9 (2011): 1617-1634.