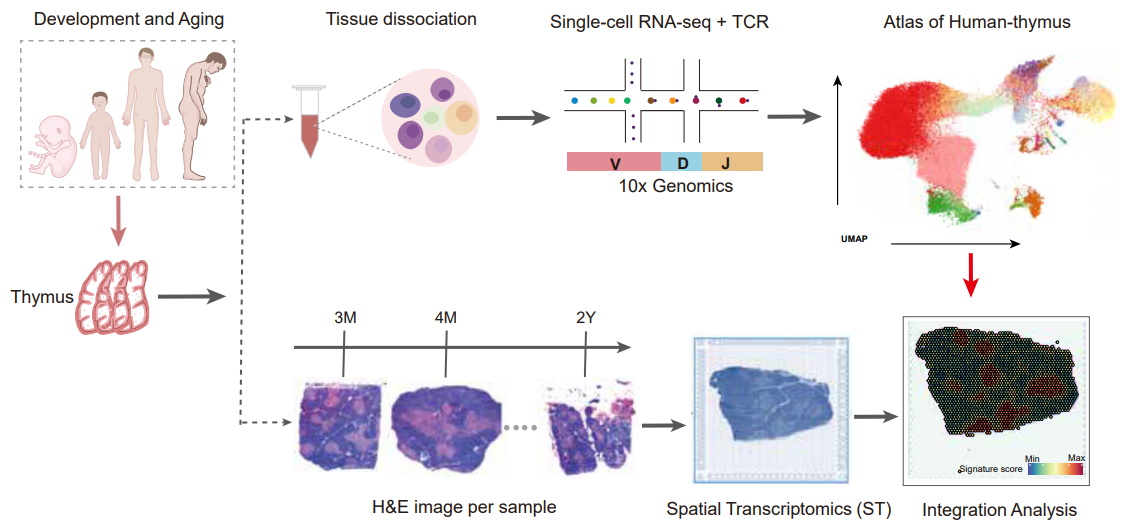
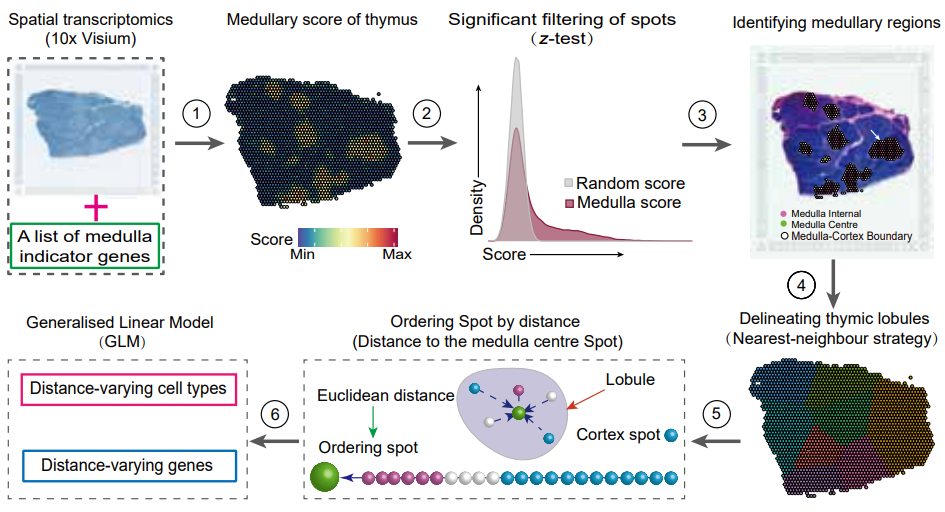
Unraveling the spatial organization and development of human thymocytes through integration of spatial transcriptomics and single-cell multi-omics profiling.
(1). All the analysis codes used in our study are provided in the source.code directory. Raw sequencing and preprocessed data can be obtained from the provided URL in our paper.
(2). 【CITATION】: Li et al. Unraveling the spatial organization and development of human thymocytes through integration of spatial transcriptomics and single-cell multi-omics profiling[J]. Nature communications, 2024.
TSO-his is a specialized tool designed for the identification of the cortex, medulla, and corticomedullary junction regions in thymic ST sections. This tool leverages statistical testing and network search algorithms to achieve its functionality.
library(devtools)
install_github("lihuamei/Thymus/thymusTSO")
library(thymusTSO)
library(dplyr)
sp.obj <- system.file('data/thymus_T2.RDS', package = 'thymusTSO') %>% readRDS
sp.obj <- tsoHis(sp.obj)
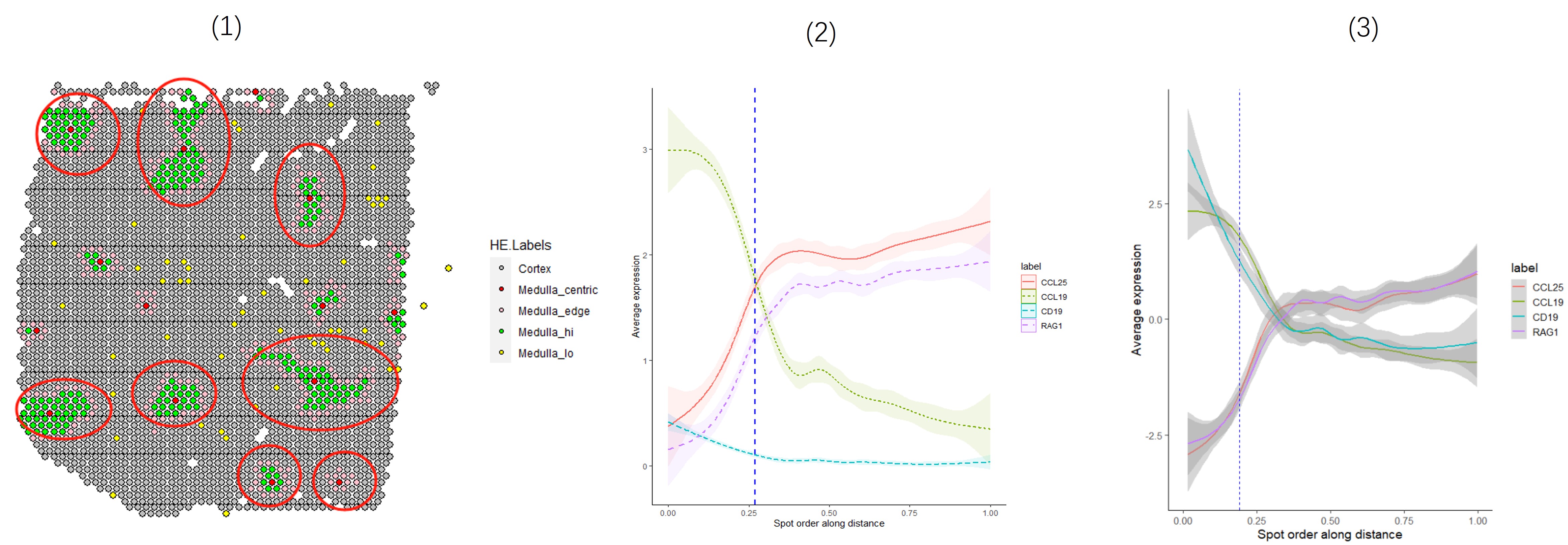
SpatialPlot(sp.obj[[1]], group.by = 'HE.Labels', cols = c('grey', 'red', 'green', 'pink', 'yellow'), pt.size.factor = 1.2) %>% `names<-`(unique(sp.obj[[1]]$HE.Labels))
fitDistLinesByGlm(sp.obj, plot.tar = c('CCL25', 'CCL19', 'CD19', 'RAG1'), degree = 4)
fitDistLinesByWindows(sp.obj, plot.tar = c('CCL25', 'CCL19', 'CD19', 'RAG1'), win = 20)
If calling trained XGBoost model for predicting cortical and medullary spots, please set call.xgb = TRUE.
sp.obj <- tsoHis(sp.obj, call.xgb = TRUE)
> sessionInfo()
R version 4.2.2 (2022-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 22000)
Matrix products: default
locale:
[1] LC_COLLATE=Chinese (Simplified)_China.utf8 LC_CTYPE=Chinese (Simplified)_China.utf8
[3] LC_MONETARY=Chinese (Simplified)_China.utf8 LC_NUMERIC=C
[5] LC_TIME=Chinese (Simplified)_China.utf8
attached base packages:
[1] splines stats graphics grDevices utils datasets methods base
other attached packages:
[1] thymusTSO_1.0.0 randomcoloR_1.1.0.1 DiagrammeR_1.0.10 tidyr_1.3.0
[5] devtools_2.4.5 usethis_2.2.2 caret_6.0-94 lattice_0.21-8
[9] ggplot2_3.4.2 Seurat_4.3.0.1 SeuratObject_5.0.1 sp_2.1-3
[13] FeatureSelection_1.0.0 verification_1.42 dtw_1.23-1 proxy_0.4-27
[17] CircStats_0.2-6 MASS_7.3-60 boot_1.3-28.1 fields_14.1
[21] viridis_0.6.3 viridisLite_0.4.2 spam_2.10-0 dplyr_1.1.0
[25] xgboost_1.7.5.1
loaded via a namespace (and not attached):
[1] utf8_1.2.3 spatstat.explore_3.2-1 reticulate_1.38.0 R.utils_2.12.2
[5] tidyselect_1.2.0 htmlwidgets_1.6.2 grid_4.2.2 ranger_0.15.1
[9] Rtsne_0.16 pROC_1.18.4 munsell_0.5.0 codetools_0.2-19
[13] ica_1.0-3 future_1.33.1 miniUI_0.1.1.1 withr_2.5.0
[17] spatstat.random_3.1-5 colorspace_2.1-0 progressr_0.14.0 knitr_1.43
[21] rstudioapi_0.15.0 stats4_4.2.2 ROCR_1.0-11 tensor_1.5
[25] listenv_0.9.1 labeling_0.4.2 polyclip_1.10-4 farver_2.1.1
[29] rprojroot_2.0.3 parallelly_1.37.1 vctrs_0.6.3 generics_0.1.3
[33] xfun_0.39 ipred_0.9-14 timechange_0.2.0 R6_2.5.1
[37] doParallel_1.0.17 spatstat.utils_3.0-3 cachem_1.0.7 promises_1.2.0.1
[41] scales_1.2.1 nnet_7.3-19 gtable_0.3.3 globals_0.16.3
[45] processx_3.8.2 goftest_1.2-3 timeDate_4022.108 rlang_1.1.1
[49] lazyeval_0.2.2 ModelMetrics_1.2.2.2 spatstat.geom_3.2-2 reshape2_1.4.4
[53] abind_1.4-5 httpuv_1.6.11 tools_4.2.2 lava_1.7.2.1
[57] ellipsis_0.3.2 RColorBrewer_1.1-3 sessioninfo_1.2.2 ggridges_0.5.4
[61] Rcpp_1.0.10 plyr_1.8.8 visNetwork_2.1.2 purrr_1.0.1
[65] ps_1.7.5 prettyunits_1.1.1 rpart_4.1.19 deldir_1.0-9
[69] pbapply_1.7-2 cowplot_1.1.1 urlchecker_1.0.1 zoo_1.8-12
[73] ggrepel_0.9.3 cluster_2.1.4 fs_1.6.2 magrittr_2.0.3
[77] data.table_1.14.8 scattermore_1.2 lmtest_0.9-40 RANN_2.6.1
[81] fitdistrplus_1.1-11 R.cache_0.16.0 matrixStats_1.0.0 pkgload_1.3.2.1
[85] patchwork_1.1.2 mime_0.12 xtable_1.8-4 gridExtra_2.3
[89] shape_1.4.6 testthat_3.1.10 compiler_4.2.2 tibble_3.2.1
[93] maps_3.4.1 V8_4.3.3 KernSmooth_2.23-22 crayon_1.5.2
[97] R.oo_1.25.0 htmltools_0.5.5 mgcv_1.9-0 later_1.3.1
[101] lubridate_1.9.2 Matrix_1.6-5 brio_1.1.3 cli_3.6.0
[105] R.methodsS3_1.8.2 parallel_4.2.2 dotCall64_1.1-1 gower_1.0.1
[109] igraph_1.5.0 pkgconfig_2.0.3 plotly_4.10.2 spatstat.sparse_3.0-2
[113] recipes_1.0.6 xml2_1.3.3 roxygen2_7.2.3 foreach_1.5.2
[117] hardhat_1.3.0 prodlim_2023.03.31 stringr_1.5.0 callr_3.7.3
[121] digest_0.6.31 sctransform_0.3.5 RcppAnnoy_0.0.21 spatstat.data_3.0-1
[125] leiden_0.4.3 uwot_0.1.16 curl_5.0.0 shiny_1.7.4.1
[129] lifecycle_1.0.4 nlme_3.1-162 jsonlite_1.8.7 desc_1.4.2
[133] fansi_1.0.4 pillar_1.9.0 fastmap_1.1.1 httr_1.4.6
[137] pkgbuild_1.4.2 survival_3.5-5 glue_1.6.2 remotes_2.4.2.1
[141] png_0.1-8 iterators_1.0.14 glmnet_4.1-7 class_7.3-22
[145] stringi_1.7.12 profvis_0.3.8 memoise_2.0.1 styler_1.10.3
[149] irlba_2.3.5.1 e1071_1.7-13 future.apply_1.11.1
Please feel free to contact us at the following email address: li_hua_mei@163.com.