Documents: http://bioinf.shenwei.me/seqkit (Usage, Tutorial, Benchmark and Development Notes)
Source code: https://github.com/shenwei356/seqkit


FASTA and FASTQ are basic and ubiquitous formats for storing nucleotide and protein sequences. Common manipulations of FASTA/Q file include converting, searching, filtering, deduplication, splitting, shuffling, and sampling. Existing tools only implement some of these manipulations, and not particularly efficiently, and some are only available for certain operating systems. Furthermore, the complicated installation process of required packages and running environments can render these programs less user friendly.
This project describes a cross-platform ultrafast comprehensive toolkit for FASTA/Q processing. SeqKit provides executable binary files for all major operating systems, including Windows, Linux, and Mac OS X, and can be directly used without any dependencies or pre-configurations. SeqKit demonstrates competitive performance in execution time and memory usage compared to similar tools. The efficiency and usability of SeqKit enable researchers to rapidly accomplish common FASTA/Q file manipulations.
- Cross-platform (Linux/Windows/Mac OS X/OpenBSD/FreeBSD, see download)
- Light weight and out-of-the-box, no dependencies, no compilation, no configuration (see download)
- UltraFast (see benchmark), multiple-CPUs supported.
- Practical functions supported by 20 subcommands (see subcommands and usage )
- Well documented (detailed usage and benchmark )
- Seamlessly parses both FASTA and FASTQ formats
- Support STDIN and gziped input/output file, easy being used in pipe
- Support custom sequence ID regular expression (especially useful for searching with ID list)
- Reproducible results (configurable rand seed in
sampleandshuffle) - Well organized source code, friendly to use and easy to extend.
| Categories | Features | seqkit | fasta_utilities | fastx_toolkit | pyfaidx | seqmagick | seqtk |
|---|---|---|---|---|---|---|---|
| Formats support | Multi-line FASTA | Yes | Yes | -- | Yes | Yes | Yes |
|FASTQ |Yes |Yes |Yes |-- |Yes |Yes
|Multi-line FASTQ |Yes |Yes |-- |-- |Yes |Yes
|Validating sequences |Yes |-- |Yes |Yes |-- |--
|Supporting RNA |Yes |Yes |-- |-- |Yes |Yes
Functions |Searching by motifs |Yes |Yes |-- |-- |Yes |-- |Sampling |Yes |-- |-- |-- |Yes |Yes |Extracting sub-sequence|Yes |Yes |-- |Yes |Yes |Yes |Removing duplicates |Yes |-- |-- |-- |Partly |-- |Splitting |Yes |Yes |-- |Partly |-- |-- |Splitting by seq |Yes |-- |Yes |Yes |-- |-- |Shuffling |Yes |-- |-- |-- |-- |-- |Sorting |Yes |Yes |-- |-- |Yes |-- |Locating motifs |Yes |-- |-- |-- |-- |-- |Common sequences |Yes |-- |-- |-- |-- |-- |Cleaning bases |Yes |Yes |Yes |Yes |-- |-- |Transcription |Yes |Yes |Yes |Yes |Yes |Yes |Translation |-- |Yes |Yes |Yes |Yes |-- |Filtering by size |Indirect|Yes |-- |Yes |Yes |-- |Renaming header |Yes |Yes |-- |-- |Yes |Yes Other features |Cross-platform |Yes |Partly |Partly |Yes |Yes |Yes |Reading STDIN |Yes |Yes |Yes |-- |Yes |Yes |Reading gzipped file |Yes |Yes |-- |-- |Yes |Yes |Writing gzip file |Yes |-- |-- |-- |Yes |--
Note 1: See version information of the softwares.
Note 2: See usage for detailed options of seqkit.
Go to Download Page for more download options and changelogs.
SeqKit is implemented in Go programming language,
executable binary files for most popular operating systems are freely available
in release page.
Just download compressed
executable file of your operating system,
and uncompress it with tar -zxvf *.tar.gz command or other tools.
And then:
-
For Linux-like systems
-
If you have root privilege simply copy it to
/usr/local/bin:sudo cp seqkit /usr/local/bin/ -
Or add the current directory of the executable file to environment variable
PATH:echo export PATH=\$PATH:\"$(pwd)\" >> ~/.bashrc source ~/.bashrc
-
-
For windows, just copy
seqkit.exetoC:\WINDOWS\system32.
For Go developer, just one command:
go get -u github.com/shenwei356/seqkit/seqkit
20 subcommands in total.
Sequence and subsequence
seqtransform sequences (revserse, complement, extract ID...)subseqget subsequences by region/gtf/bed, including flanking sequencesslidingsliding sequences, circular genome supportedstatsimple statistics of FASTA filesfaidxcreate FASTA index file
Format conversion
fx2tabcovert FASTA/Q to tabular format (and length/GC content/GC skew)tab2fxcovert tabular format to FASTA/Q formatfq2facovert FASTQ to FASTA
Searching
grepsearch sequences by pattern(s) of name or sequence motifslocatelocate subsequences/motifs
Set operations
rmdupremove duplicated sequences by id/name/sequencecommonfind common sequences of multiple files by id/name/sequencesplitsplit sequences into files by id/seq region/size/partssamplesample sequences by number or proportionheadprint first N FASTA/Q records
Edit
replacereplace name/sequence by regular expressionrenamerename duplicated IDs
Ordering
shuffleshuffle sequencessortsort sequences by id/name/sequence
Misc
versionprint version information and check for update
SeqKit uses author's lightweight and high-performance bioinformatics packages bio for FASTA/Q parsing, which has high performance close to the famous C lib klib (kseq.h).
SeqKit seamlessly support FASTA and FASTQ format.
Sequence format is automatically detected.
All subcommands except for faidx can handle both formats.
And only when some commands (subseq, split, sort and shuffle)
which utilise FASTA index to improve perfrmance for large files in two pass mode
(by flag --two-pass), only FASTA format is supported.
Sequence type (DNA/RNA/Protein) is automatically detected by leading subsequences
of the first sequences in file or STDIN. The length of the leading subsequences
is configurable by global flag --alphabet-guess-seq-length with default value
of 10000. If length of the sequences is less than that, whole sequences will
be checked.
By default, most softwares, including seqkit, take the leading non-space
letters as sequence identifier (ID). For example,
| FASTA header | ID |
|---|---|
| >123456 gene name | 123456 |
| >longname | longname |
| >gi|110645304|ref|NC_002516.2| Pseudomona | gi|110645304|ref|NC_002516.2| |
But for some sequences from NCBI,
e.g. >gi|110645304|ref|NC_002516.2| Pseudomona, the ID is NC_002516.2.
In this case, we could set sequence ID parsing regular expression by global flag
--id-regexp "\|([^\|]+)\| " or just use flag --id-ncbi. If you want
the gi number, then use --id-regexp "^gi\|([^\|]+)\|".
For some commands, including subseq, split, sort and shuffle,
when input files are (plain or gzipped) FASTA files,
FASTA index would be optional used for
rapid access of sequences and reducing memory occupation.
ATTENTION: the .seqkit.fai file created by SeqKit is slightly different from .fai file
created by samtools. SeqKit uses full sequence head instead of just ID as key.
The validation of sequences bases and complement process of sequences are parallelized for large sequences.
Parsing of line-based files, including BED/GFF file and ID list file are also parallelized.
The Parallelization is implemented by multiple goroutines in golang
which are similar to but much
lighter weight than threads. The concurrency number is configurable with global
flag -j or --threads (default value: 1 for single-CPU PC, 2 for others).
Most of the subcommands do not read whole FASTA/Q records in to memory,
including stat, fq2fa, fx2tab, tab2fx, grep, locate, replace,
seq, sliding, subseq.
Note that when using subseq --gtf | --bed, if the GTF/BED files are too
big, the memory usage will increase.
You could use --chr to specify chromesomes and --feature to limit features.
Some subcommands need to store sequences or heads in memory, but there are
strategy to reduce memory occupation, including rmdup and common.
When comparing with sequences, MD5 digest could be used to replace sequence by
flag -m (--md5).
Some subcommands could either read all records or read the files twice by flag
-2 (--two-pass), including sample, split, shuffle and sort.
They use FASTA index for rapid acccess of sequences and reducing memory occupation.
Subcommands sample and shuffle use random function, random seed could be
given by flag -s (--rand-seed). This makes sure that sampling result could be
reproduced in different environments with same random seed.
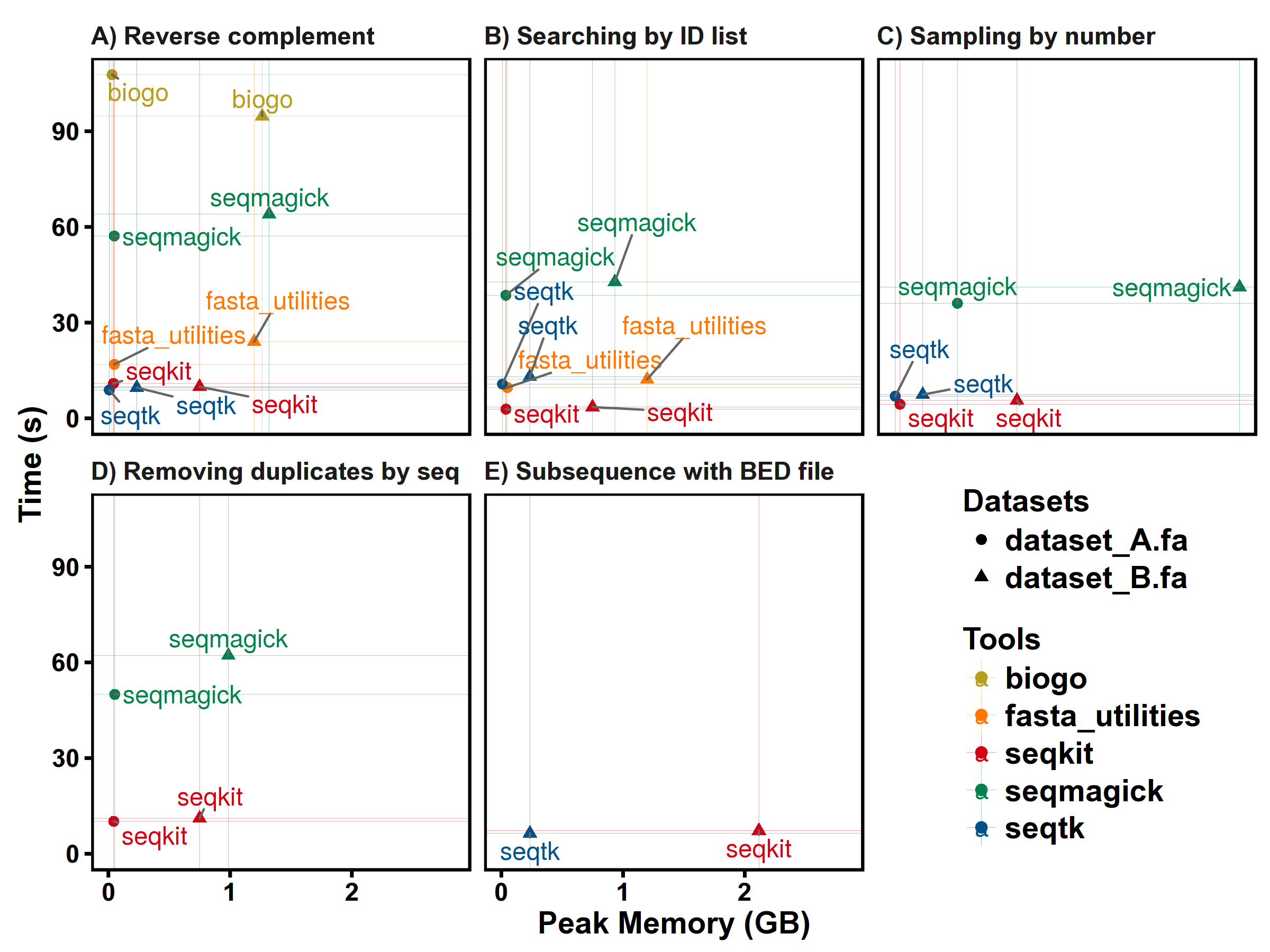
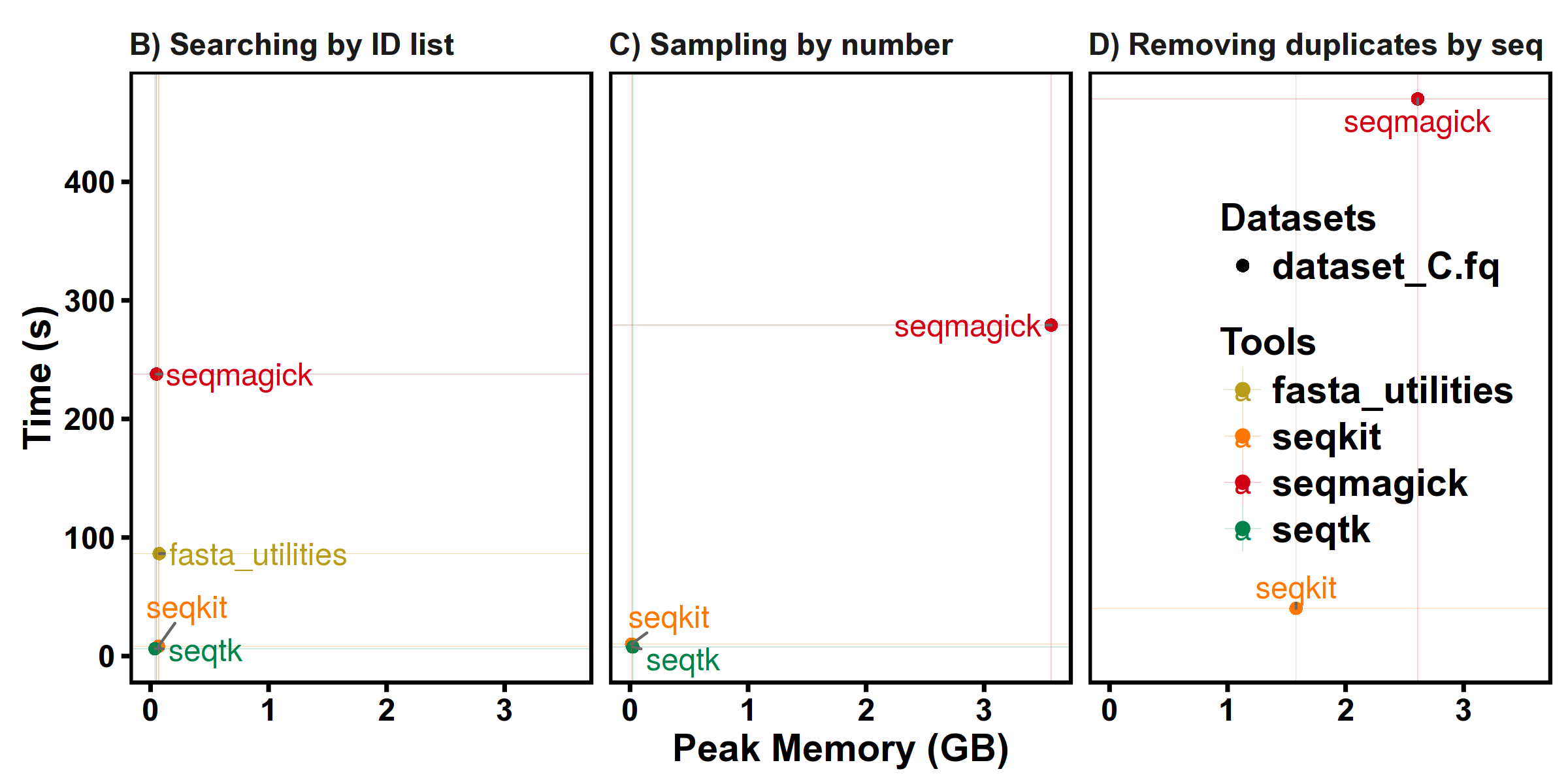
More details: http://bioinf.shenwei.me/seqkit/benchmark/
Datasets:
$ seqkit stat *.fa
file format type num_seqs sum_len min_len avg_len max_len
dataset_A.fa FASTA DNA 67,748 2,807,643,808 56 41,442.5 5,976,145
dataset_B.fa FASTA DNA 194 3,099,750,718 970 15,978,096.5 248,956,422
dataset_C.fq FASTQ DNA 9,186,045 918,604,500 100 100 100
SeqKit version: v0.3.1.1
FASTA:
FASTQ:
W Shen, S Le, Y Li*, F Hu*. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLOS ONE. doi:10.1371/journal.pone.0163962.
We thank Lei Zhang for testing of SeqKit, and also thank Jim Hester, author of fasta_utilities, for advice on early performance improvements of for FASTA parsing and Brian Bushnell, author of BBMaps, for advice on naming SeqKit and adding accuracy evaluation in benchmarks. We also thank Nicholas C. Wu from the Scripps Research Institute, USA for commenting on the manuscript and Guangchuang Yu from State Key Laboratory of Emerging Infectious Diseases, The University of Hong Kong, HK for advice on the manuscript.
We thank Li Peng for reporting many bugs.
Email me for any problem when using seqkit. shenwei356(at)gmail.com
Create an issue to report bugs, propose new functions or ask for help.