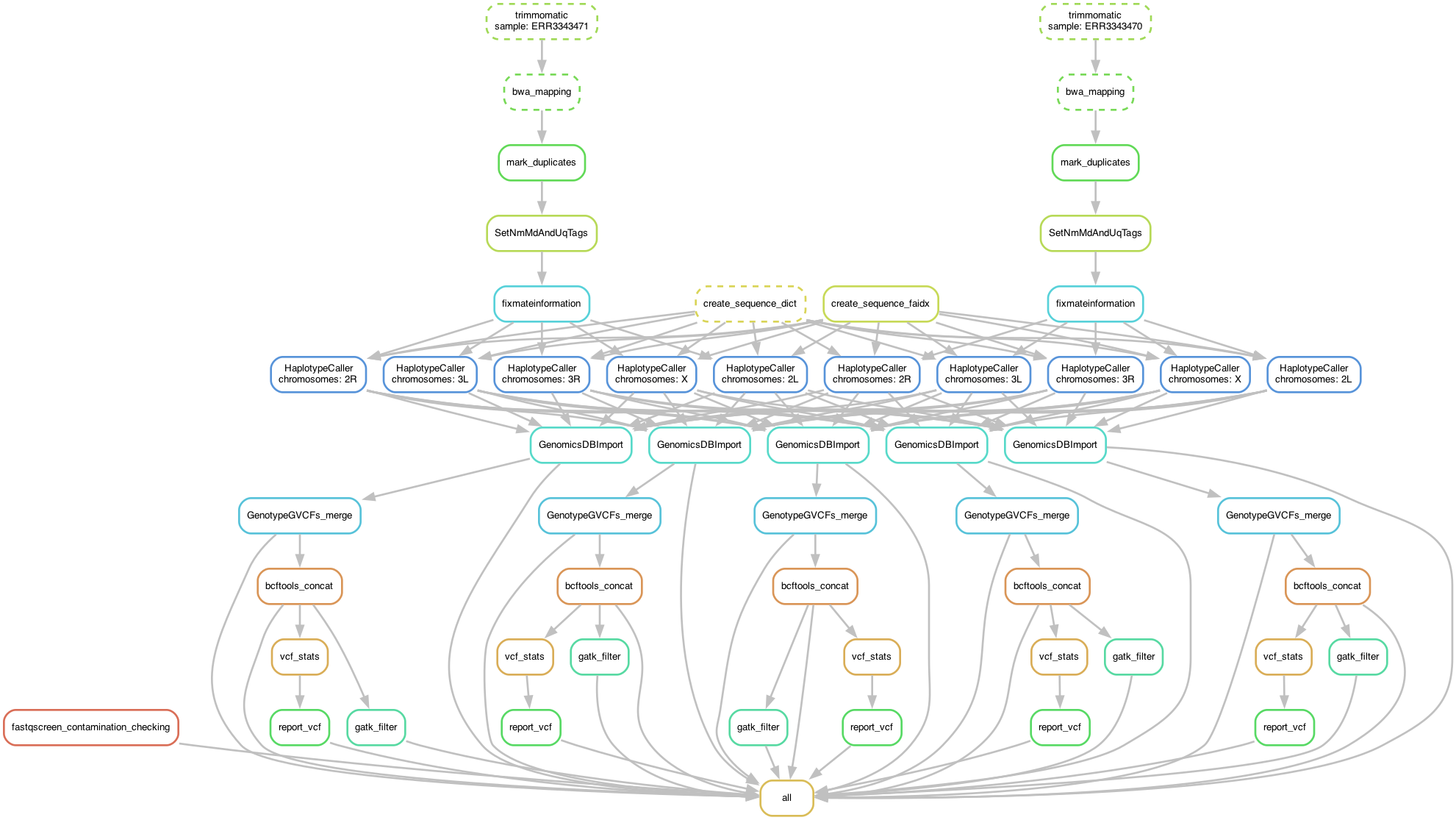
SHAVE2: SHort-read Alignment pipeline for VEctor v.2 - with HaplotypeCaller variant caller - SLURM version
SHAVE2 is a bioinformatic pipeline used for mosquitoes (Aedes / Anopheles) genome alignments from Illumina short reads, based on GATK Best Practices, except for the BQSR and VQSR steps.
In brief, SHAVE2 remove adatpers, report quality reads, aligns reads to a reference genome, fix incorrect mates, mark duplicates, add indel qualities to BAM files and call variants and genotypes.
Indel realignment was dropped by Broad Institute about three years ago, as they found that this step was no longer useful when the variant calling was done with HaplotypeCaller or Mutect2, which implement a more sophisticated and effective form of realignment. see here. SHAVE2 use HaplotypeCaller in ERC mode to call variants and obtain the genotype likelihoods. Then, GenomicsDBImport import single-sample GVCFs into GenomicsDB before GenotypeGVCFs perform a joint genotyping on samples pre-called with HaplotypeCaller. The last major step is a fully customizable hard-filtering, using GATK VariantFiltration instead of Variant Qualtity Score Recalibration (VQSR).
Base Quality Score Recalibration step needs as input a known variation VCF file, refering to the Ensembl-Variation database or dbSNP database who stores areas of genome that differ between individual genomes ("variants"). However, we do not have any prior list of known variants for our mosquito species, that's why we cannot do BQSR.
But the value of BQSR is increasingly being questioned as mappers and callers are typically updated. As an example: using HaplotypeCaller instead of UnifiedGenotyper greatly improves the handling of indels.
Variant Quality Score recalibration is probably the hardest part of the Best Practices to get right according to Broad Institute. In a nutshell, it is a sophisticated filtering technique applied on the variant callset that uses machine learning to model the technical profile of variants in a training set and uses that to filter out probable artifacts from the callset.
The key point is that it use known, highly validated variant resources (omni, 1000 Genomes, hapmap) to select a subset of variants within our callset that we’re really confident are probably true positives (the training set). Unfortunately, no highly validated variant resource is available for mosquitoes at this time, so we decided to apply hard-filtering and leave the choice of parameters to the user.
Written for MOVE-ADAPT project.
- Control reads quality (multiQC html report) and clean it
- Align reads (bam files),
- Mark duplicates to BAM files,
- Add MD and NM tag to BAM files,
- Fix mates,
- BAM file validation according to SAM/BAM specifications,
- Variants calling (vcf files),
- Genotyping,
- VCF compression,
- VCF indexing
V2.2023.07.13
Clone (HTTPS) the SHAVE2 repository on github:
git clone git@github.com:ltalignani/SHAVE2.git
Difference between Download and Clone:
- To create a copy of a remote repository’s files on your computer, you can either Download or Clone the repository
- If you download it, you cannot sync the repository with the remote repository on GitHub
- Cloning a repository is the same as downloading, except it preserves the Git connection with the remote repository
- You can then modify the files locally and upload the changes to the remote repository on GitHub
- You can then update the files locally and download the changes from the remote repository on GitHub
git pull --verboseThen, upload the SHAVE2/ archive on your slurm cluster.
- Copy your paired-end reads in .fastq.gz format files into: .raw/ directory
- Execute the sbatch Start_shave2_slurm.sh bash script to run the SHAVE2 pipeline
sbatch Start_shave2_slurm.shYours analyzes will start with default configuration settings
Option-1: Edit config.yaml file in ./config/ directory
Option-2: Edit fastq-screen.conf file in ./config/ directory
Yours results are available in ./results/ directory, as follow:
(file names keep track which tools / params was used: <sample>_<aligner>_<mincov>)
| File | Object |
|---|---|
| fastq-screen | raw reads putative contaminations reports for each samples, in html, png and txt formats |
| fastqc | raw reads quality reports for each samples, in html and zip formats |
| multiqc | fastq-screen and fastqc results agregation report for all samples, in html format |
| qualimap | facilitate the quality control of alignment sequencing data and its derivatives like feature counts, in html format |
| validatesamfile | This tool reports on the validity of a SAM or BAM file relative to the SAM format specification, in txt format |
| File | Object |
|---|---|
| trimmomatic/ directory | paired reads, without adapters and quality trimmed, in fastq.gz format |
| File | Object |
|---|---|
| mark-dup.bam | read alignments, in bam format (can be visualized in, i.e. IGV) |
| mark-dup.bai | bam indexes bai use in i.e. IGV with ./resources/genomes/AalbF3.fasta |
| markdup_metrics.txt | bam metrics created by Picard MarkDuplicates |
| File | Object |
|---|---|
| md_fixed.bam | polished bam files , in bam format |
| md_fixed.bam.bai | polished bam indexes, in bai format |
| File | Object |
|---|---|
| variant-call.g.vcf | SNVs and Indels calling in gvcf format |
| variant-call.g.vcf.idx | SNV and Indels calling in idx format |
| genomicdb | g.vcfs merged by GenomicsDBImport for GenotypeGVCFs |
| genotyped.vcf | SNVs genotyped in vcf format |
| genotyped.vcf.idx | genotype index in idx format (automatically generated by GenotypeGVCFs) |
| combinedGVCF.vcf.gz | alternative to GenomicsDBImport, is a merging of GVCFs that can eventually be input into GenoTypeGVCFs |
| GenotypeGVCFs.hf.vcf.gz | genotyped SNVs hard-filtered, in vcf format (default config: tempdir, removed, save disk usage)_ |
| File | Object |
|---|---|
| dag | directed acyclic graph of jobs, in pdf and png formats |
| rulegraph | dependency graph of rules, in pdf and png formats (less crowded than above DAG of jobs, but also show less information) |
| filegraph | dependency graph of rules with their input and output files in the dot language, in pdf and png formats (an intermediate solution between above DAG of jobs and the rule graph) |
- All non-empty log for each tool and each sample
- files_summary.txt: summary of all files created by the workflow, in txt format
(columns: filename, modification time, rule version, status, plan)
If needed, see or edit default settings in config.yaml file in ./config/ directory
samples and units aren't necessary for the moment (update in progress)
Modules that will be loaded for each tool. Change the path to correspond to your cluster.
- ref_name: reference name used for genome mapping (default config: 'Anopheles-gambiae-PEST_CHROMOSOMES_AgamP4')
- path: path to genome reference
- reference: reference sequence path used for genome mapping (default config: 'AGAMP4')
- index: reference sequence index used for genome mapping (default config: 'Anopheles-gambiae-PEST_CHROMOSOMES_AgamP4.fa.fai')
- dictionary: reference sequence dictionary used by GATK's tools (default config: 'Anopheles-gambiae-PEST_CHROMOSOMES_AgamP4.dict')
- tmpdir: temporary directory (default config: '$TMPDIR')
- genomicsdbimport: genomicsdbimport flags (default config: "")
- genotypegvcfs: genotypegvcfs flags (default config: '-include-non-variant-sites')
- haplotypecaller: haplotypecaller flags (default config: '-ERC GVCF --output-mode EMIT_ALL_CONFIDENT_SITES')
- alleles_target: path to VCF containing known varition sites (default config: "--alleles xxxx"). Replace "xxxx" by path to the VCF containing known variation site.
- path: path the BWA indexes (default config: 'resources/indexes/bwa/')
- config: path to the fastq-screen configuration file (default config: ./config/fastq-screen.conf)
- subset: do not use the whole sequence file, but create a temporary dataset of this specified number of read (default config: '1000')
- aligner: specify the aligner to use for the mapping. Valid arguments are 'bowtie', bowtie2' or 'bwa' (default config: 'bwa')
- vqsr: not coded at this time
- hard: settings for VCFs hardfiltering (default config:myfilter": "QD < 2.0 || FS > 60.0 || MQ < 40.0 || MQRankSum < -12.5 || ReadPosRankSum < -8.0)
- BAM: Binary Alignment Map
- BAI: BAM Indexes
- FASTA: Fast-All
- FASTQ: FASTA with Quality
- FAI: FASTA Indexes
- SAM: Sequence Alignment Map
🖥️️ Start_shave2_slurm.sh
📚 README.md
📚 LICENSE
📂 Cluster_logs/
📂 visuals/
└── 📈 DAG.png
📂 config/
├── ⚙️ config.yaml
└── ⚙️ fastq-screen.conf
📂 raw/
│ ├── 🛡️ .gitkeep
│ ├── 📦 ERR3343471_R1.fastq.gz
│ └── 📦 ERR3343471_R2.fastq.gz
📂 resources/
├── 📂 genomes/
│ └── 🧬 AalbF3.fasta
├── 📂 indexes/
│ ├── 📂 bwa/
│ │ └── 🗂️ AGAMP4
├── 📂 Adapters
│ ├── 🧬 nextera
│ ├── 🧬 truseq2-pe
│ ├── 🧬 truseq2-se
│ ├── 🧬 truseq3-pe
│ ├── 🧬 truseq3-pe2
│ └── 🧬 truseq3-se
│
├── 📂 results/
├── 📂 slurm/
│ ├── ⚙️ config.yaml
│ └── 🖥️️ status-sacct.sh
📂 workflow/
├── 📂 envs/
│ ├── 🍜 bcftools-1.14.yaml
│ ├── 🍜 bedtools-2.30.0.yaml
│ ├── 🍜 bowtie2-2.4.4.yaml
│ ├── 🍜 bwa-0.7.17.yaml
│ ├── 🍜 cutadapt-3.5.yaml
│ ├── 🍜 fastq-screen-0.14.0.yam
│ ├── 🍜 fastqc-0.11.9.yaml
│ ├── 🍜 gatk-3.8.yaml
│ ├── 🍜 gatk-4.3.0.0.yaml
│ ├── 🍜 gawk-5.1.0.yaml
│ ├── 🍜 lofreq-2.1.5.yaml
│ ├── 🍜 multiqc-1.11.yaml
│ ├── 🍜 picard-2.27.4.yaml
│ ├── 🍜 samtools-1.14.yaml
│ └── 🍜 sickle-trim-1.33.yaml
├── 📂 report/
├── 📂 rules/
│ ├── 📜 calling.smk
│ ├── 📜 common.smk
│ ├── 📜 filtering.smk
│ ├── 📜 mapping.smk
│ ├── 📜 polishing.smk
│ ├── 📜 stats.smk
│ └── 📜 vcf_stats.smk
├── 📂 schemas/
├── 📂 scripts/
│ ├── 🍜 report_vcf.rmd
│ └── 🍜 report.rmd
└── 📜 snakefile.smk- Read The Fabulous Manual!
- Read de Awsome Wiki! (todo...)
- Create a new issue: Issues > New issue > Describe your issue
- Send an email to loic.talignani@ird.fr
- Add a wiki!
- Loïc TALIGNANI (Developer and Maintener)
- I would like to thanks Sebastien Ravel, CIRAD ingeneer, for his great help for the variant calling part.
Open to contributions!
Testing code, finding issues, asking for update, proposing new features...
Use Git tools to share!
This project is regularly updated and actively maintened
However, you can be volunteer to step in as developer or maintainer
For information about main git roles:
- Guests are not active contributors in private projects, they can only see, and leave comments and issues
- Reporters are read-only contributors, they can't write to the repository, but can on issues
- Developers are direct contributors, they have access to everything to go from idea to production
Unless something has been explicitly restricted - Maintainers are super-developers, they are able to push to master, deploy to production
This role is often held by maintainers and engineering managers - Owners are essentially group-admins, they can give access to groups and have destructive capabilities
Sustainable data analysis with Snakemake
Felix Mölder, Kim Philipp Jablonski, Brice Letcher, Michael B. Hall, Christopher H. Tomkins-Tinch, Vanessa Sochat, Jan Forster, Soohyun Lee, Sven O. Twardziok, Alexander Kanitz, Andreas Wilm, Manuel Holtgrewe, Sven Rahmann, Sven Nahnsen, Johannes Köster
F1000Research (2021)
DOI: https://doi.org/10.12688/f1000research.29032.2
Publication: https://f1000research.com/articles/10-33/v1
Source code: https://github.com/snakemake/snakemake
Documentation: https://snakemake.readthedocs.io/en/stable/index.html
Tabix: fast retrieval of sequence features from generic TAB-delimited files
Heng Li
Bioinformatics, Volume 27, Issue 5 (2011)
DOI: https://doi.org/10.1093/bioinformatics/btq671
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3042176/
Source code: https://github.com/samtools/samtools
Documentation: http://samtools.sourceforge.net
GATK: A MapReduce framework for analyzing next-generation DNA sequencing data Genome Research, Volume 20: 1297-1303 (2010) DOI: https://doi.org/10.1101/gr.107524.110 Publication: https://genome.cshlp.org/content/20/9/1297 Source code:https://github.com/broadinstitute/gatk Documentation:https://gatk.broadinstitute.org/hc/en-us
**Picard-tools: ** Broad Institute, GitHub repository (2019) DOI: Publication: Source code:https://github.com/broadinstitute/picard](https://github.com/broadinstitute/picard) Documentation:https://broadinstitute.github.io/picard/
The AWK Programming Language
Al Aho, Brian Kernighan and Peter Weinberger
Addison-Wesley (1988)
ISBN: https://www.biblio.com/9780201079814
Publication:
Source code: https://github.com/onetrueawk/awk
Documentation: https://www.gnu.org/software/gawk/manual/gawk.html
Twelve years of SAMtools and BCFtools
Petr Danecek, James K Bonfield, Jennifer Liddle, John Marshall, Valeriu Ohan, Martin O Pollard, Andrew Whitwham, Thomas Keane, Shane A McCarthy, Robert M Davies and Heng Li
GigaScience, Volume 10, Issue 2 (2021)
DOI: https://doi.org/10.1093/gigascience/giab008
Publication: https://academic.oup.com/gigascience/article/10/2/giab008/6137722
Source code: https://github.com/samtools/samtools
Documentation: http://samtools.sourceforge.net
Fast and accurate short read alignment with Burrows-Wheeler Transform
Heng Li and Richard Durbin
Bioinformatics, Volume 25, Aricle 1754-60 (2009)
DOI: https://doi.org/10.1093/bioinformatics/btp324
Publication: https://pubmed.ncbi.nlm.nih.gov/19451168@
Source code: https://github.com/lh3/bwa
Documentation: http://bio-bwa.sourceforge.net
Trimmomatic: A sliding-window, adaptive, quality-based trimming tool for FastQ files
Joshi NA and Fass JN
_(2011)
DOI: https://doi.org/10.1093/bioinformatics/btu170
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4103590/pdf/btu170.pdf
Source code: https://github.com/usadellab/Trimmomatic
Documentation: https://github.com/usadellab/Trimmomatic
MultiQC: summarize analysis results for multiple tools and samples in a single report
Philip Ewels, Måns Magnusson, Sverker Lundin and Max Käller
Bioinformatics, Volume 32, Issue 19 (2016)
DOI: https://doi.org/10.1093/bioinformatics/btw354
Publication: https://academic.oup.com/bioinformatics/article/32/19/3047/2196507
Source code: https://github.com/ewels/MultiQC
Documentation: https://multiqc.info
FastQ Screen: A tool for multi-genome mapping and quality control
Wingett SW and Andrews S
F1000Research (2018)
DOI: https://doi.org/10.12688/f1000research.15931.2
Publication: https://f1000research.com/articles/7-1338/v2
Source code: https://github.com/StevenWingett/FastQ-Screen
Documentation: https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen
FastQC: A quality control tool for high throughput sequence data
Simon Andrews
Online (2010)
DOI: https://doi.org/
Publication:
Source code: https://github.com/s-andrews/FastQC
Documentation: https://www.bioinformatics.babraham.ac.uk/projects/fastqc