The docker installation procedure should take less than 15 minutes.
First, make sure that docker is installed on your system. You can find instructions on how to install docker here. To make sure that the installation was successful, open a terminal window and enter:
docker ps
Clone, you need to clone the github repo:
git clone https://github.com/ludvigla/RRST
Once the RRST GitHub repo has been downloaded, we can navigate into the newly created folder.
cd RRST
Now we can generate a container from the ludlar/rrst image. This image contains an environment
with an installation of R and all R packages necessary to run the analyses.
Note that you need to be in the correct folder (i.e. the RRST folder cloned from GitHub). You can change
--memory flag as you see fit, but the code has only been tested with 16GiB of RAM.
You can provide your own password for the RStudio server by replacing YOURPASSWORD, for example
-e PASSWORD=12345678.
Now run the code below (don't forget to replace YOURPASSWORD) in the terminal:
sudo docker run -d -p 1337:8787 --name RRST -e PASSWORD=YOURPASWORD --memory=16g --mount type=bind,source="$(pwd)",target=/home/rstudio -e ROOT=TRUE ludlar/rrst:latest
If you run the code with sudo, you will need to enter your computer password; however, it might work well without sudo.
When the docker run command is finished, you can check that the container is running by typing:
docker container ls -a
Now you can go open a browser (e.g. Chrome) and enter: localhost:1337 in the address bar.
This should take you to a login page for an RStudio server where you can enter the user name and your password to login. Please follow the instruction below (Instruction for use) more more information on how to get started with the analysis.
- user name: rstudio
- password:
YOURPASSWORD
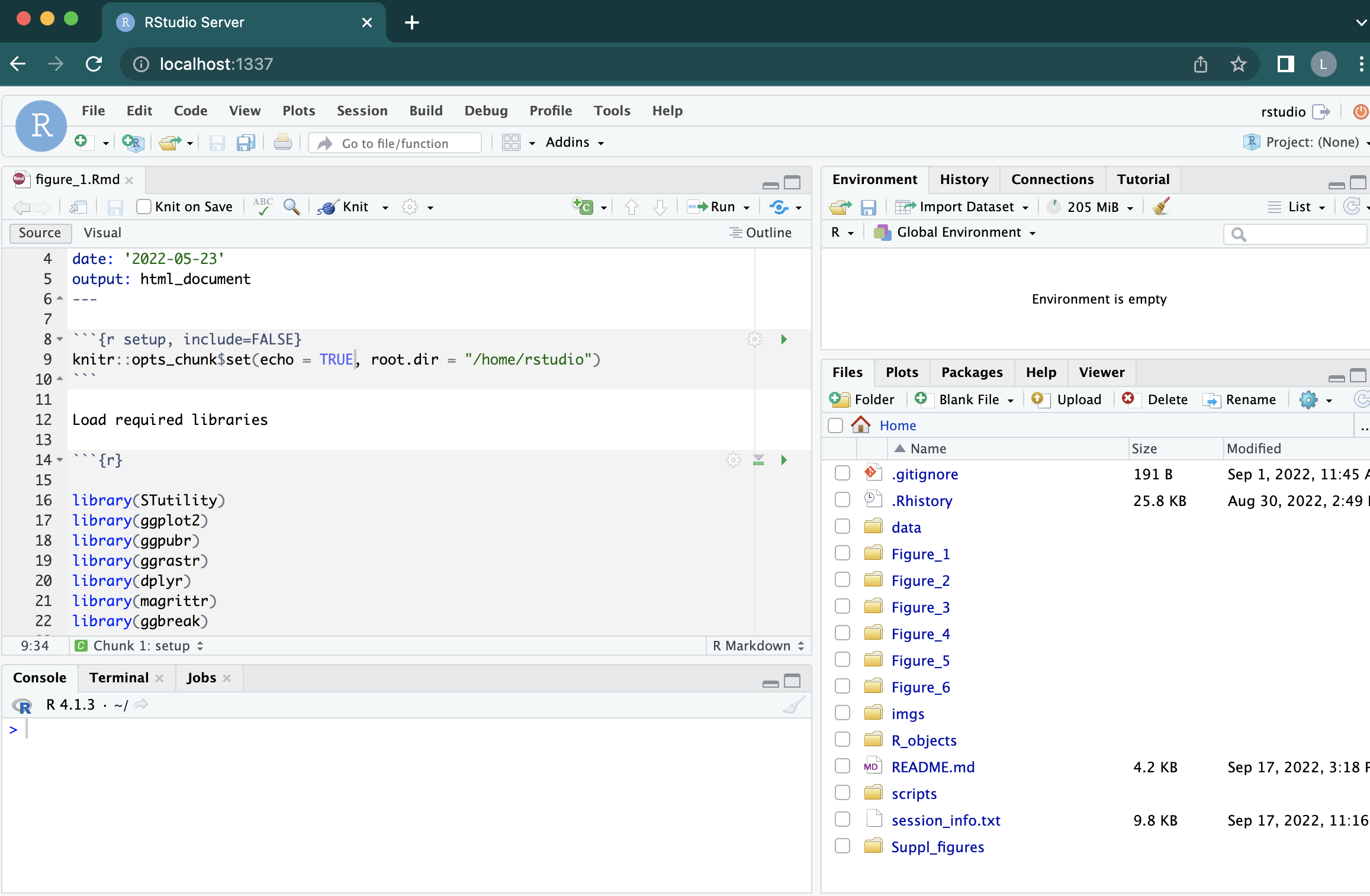
When you have logged in, you should see something like this:
You can start and stop the container by running the following commands from the terminal:
docker start RRST
docker stop RRST
If you run out of memory in the container, check if you can allocate more with
docker run --memory=<SOME VALUE>.
You might also want to increase the memory limit in docker. If you run docker info you can check how much memory is
available to docker, e.g. "Total memory: 8GiB". If you have more resources available on your system, you can increase
this number. If you are using the Docker desktop app, open it and go to Preferences -> Resources and increase the memory.
The docker memory limit puts a limitation on the memory available to the container.
For example, if you try to set --memory=16g when the the docker memory limit is 8GiB, you will end up with 8GiB.
You can get more info about how to manage resources here on the docker website.
Together, all analyses should take less than an hour to run on a laptop with specs comparable to those specified under System requirements below.
When you have opened the rstudio server, you should see one folder for each main figue in the
file viewer pane (bottom right). Inside each of these folders, there is an .Rmd notebook
with instructions that you can run to reproduce the analyses and generate plots used
in the manuscript.
The data needed to run the analyses is stored at Mendeley Data. Instructions on how to
download this data can be found in the beginning of each .Rmd notebook.
Supplementary figures are produced in the same .Rmd notebooks. For example, supplementary
figures related to Figure 1 can be produced in the figure_1.Rmd notebook.
All main figure plots exported in the notebooks are available in the plots/ sub folders inside
each figure folder. Supplementary figures are located in the Suppl_figures/folder. The expected
output (plots) from the notebooks should be comparable to these exported figures and the figures
in the manuscript.
The code has been tested on a Macbook Pro (2017) with the specs below:
* Processor : 3.1 GHz Quad-Core Intel Core i7
* Memory : 16 GB 2133 MHz LPDDR3
* Graphics : Intel HD Graphics 630 1536 MB
You can find details about the R environment used in the session_info.txt file. We recommend
using the docker image to run the analyses in a reproducible R environment. Using more recent
installations of certain R packages may lead to slightly different results, in particular
for analyses that use functions from the R packages Seurat and sctransform.