"Detection of tumour infiltrating lymphocytes in CD3 and CD8 stained histopathological images using a two-phase deep CNN"
The research article is available in Photodiagnosis and Photodynamic Therapy Journal, Volume 37, and can be accessed using this Link
- Muhammad Mohsin Zafar
- Zunaira Rauf
- Anabia Sohail
- Abdul Rehman Khan
- Muhammad Obaidullah
- Saddam Hussain Khan
- Yeon Soo Lee
- Asifullah Khan
This GitHub repository is released along with our submission "Detection of Lymphocytes in Histopathological Images using a Two-Phase Deep CNN" in Photodiagnosis and Photodynamic Therapy Journal. The repository at the moment contains only the inference code. Later on after the acceptance of our article for publication we intend on releasing complete code that shall include code for multiple experiments and training the models on same parameters as we did.
Following are the prerequisites for using this repository.
- Python >= 3.7
All modules that were used, are mentioned in requirements.txt file.
gdown==3.13.0
opencv-python~=4.5.1.48
numpy~=1.19.2
matplotlib==3.3.4
tqdm~=4.59.0
pillow~=8.1.2
scikit-image==0.17.2
pytorch==1.8.0
torchvision==0.9.0
detectron2==0.5
Please follow the instructions in order to use this repository effectively
git clone https://github.com/m-mohsin-zafar/tdc-lcFor installing PyTorch you can visit their website here OR
OSX
# conda
conda install pytorch==1.8.0 torchvision==0.9.0 torchaudio==0.8.0 -c pytorchLinux and Windows
# CUDA 10.2
conda install pytorch==1.8.0 torchvision==0.9.0 torchaudio==0.8.0 cudatoolkit=10.2 -c pytorch
# CUDA 11.1
conda install pytorch==1.8.0 torchvision==0.9.0 torchaudio==0.8.0 cudatoolkit=11.1 -c pytorch -c conda-forge
# CPU Only
conda install pytorch==1.8.0 torchvision==0.9.0 torchaudio==0.8.0 cpuonly -c pytorchOSX
pip install torch==1.8.0 torchvision==0.9.0 torchaudio==0.8.0Linux and Windows
# RocM 4.0.1 (Linux only)
pip install torch -f https://download.pytorch.org/whl/rocm4.0.1/torch_stable.html
pip install ninja
pip install 'git+https://github.com/pytorch/vision.git@v0.9.0'
# CUDA 11.1
pip install torch==1.8.0+cu111 torchvision==0.9.0+cu111 torchaudio==0.8.0 -f https://download.pytorch.org/whl/torch_stable.html
# CUDA 10.2
pip install torch==1.8.0 torchvision==0.9.0 torchaudio==0.8.0
# CPU only
pip install torch==1.8.0+cpu torchvision==0.9.0+cpu torchaudio==0.8.0 -f https://download.pytorch.org/whl/torch_stable.htmlOfficial Documentation for installing the Detectron2 API can be found here.
One method for installation of Detectron2 is as follows:
python -m pip install 'git+https://github.com/facebookresearch/detectron2.git'
# (add --user if you don't have permission)
# Or, to install it from a local clone:
git clone https://github.com/facebookresearch/detectron2.git
python -m pip install -e detectron2
# On macOS, you may need to prepend the above commands with a few environment variables:
CC=clang CXX=clang++ ARCHFLAGS="-arch x86_64" python -m pip install ...Finally, install rest of the modules from requirements.txt
pip install -r requirements.txtconda install --file requirements.txtFor a better understanding of repository please follow along with the explanation below.
.
├── checkpoints # Contains all the weight files of model
│ ├── download.py
│ └── README.md
├── data # Input image patches that you want to perform inference on
│ ├── test_10080.png
│ ├── test_10227.png
├── main.py # Entry Point file, you just need to run this
├── models # Code for building the models is placed here
│ ├── build.py
│ ├── configs.py
│ └── __init__.py
├── output # Output of inference can be found here
│ ├── result_count.csv # The file contains count of lymphocytes found against each input image
│ └── viz # Ouput visuals will be save in this folder
├── pipeline # Code for complete inference pipeline
│ ├── inference.py
│ ├── __init__.py
│ ├── postprocess.py
│ ├── preprocess.py
│ └── tmp # Temporary DAB images that are saved during inference
├── README.md
└── requirements.txtFollowing are the steps to run inference.
Assuming, you are already inside the repository folder.
# Change pwd to ./checkpoints
cd checkpoints
# Run download.py file
python download.pyAfter running this, you'll see that two *.pth files are downloaded in checkpoints folder
Place your data of Lymphocyte Image Patches in data folder. Preferably, the input images should be *.png format.
OR
You may use the already placed images in the folder just for testing purposes.
Few sample images are shown below.
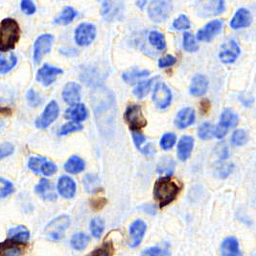
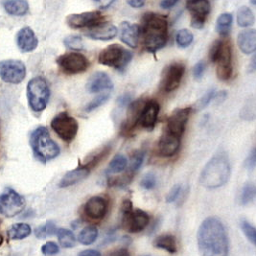
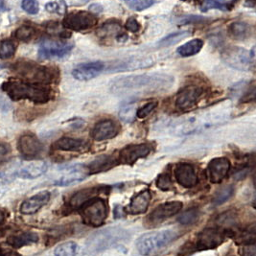
Assuming, you are in the root repository folder. You just need to execute main.py file.
python main.pyThe output is saved in the output folder. Inside the folder you will find a csv file named, "result_count.csv" which contains two columns as per following pattern.
| file_name | count |
|---|---|
| test_15907.png | 0 |
| test_1949.png | 3 |
| test_12565.png | 12 |
Also, in the output folder you'll find another folder named "viz" which shall contain the output visualizations. You may find some output visualizations to be plain black images, this would occur in case there are no predicted lymphocytes i.e. count=0. Some sample output visualizations are shown below.
We are thankful to Pakistan Institute of Engineering and Applied Sciences, Islamabad (PIEAS) and Pattern Recognition Lab for their kind support and facilitation in computational resources.
If you use the code in your research or wish to refer to the baseline results published in the article, please use the following BibTeX entry.
@article{ZAFAR2022102676,
title = {Detection of tumour infiltrating lymphocytes in CD3 and CD8 stained histopathological images using a two-phase deep CNN},
journal = {Photodiagnosis and Photodynamic Therapy},
volume = {37},
pages = {102676},
year = {2022},
issn = {1572-1000},
doi = {https://doi.org/10.1016/j.pdpdt.2021.102676},
url = {https://www.sciencedirect.com/science/article/pii/S1572100021004932},
author = {Muhammad Mohsin Zafar and Zunaira Rauf and Anabia Sohail and Abdul Rehman Khan and Muhammad Obaidullah and Saddam Hussain Khan and Yeon Soo Lee and Asifullah Khan},
keywords = {tumor-infiltrating Lymphocytes (TILs), Mask R-CNN, Histopathological images, Lymphocyte detection, Deep Convolutional Neural Network (DCNN)}
}```


