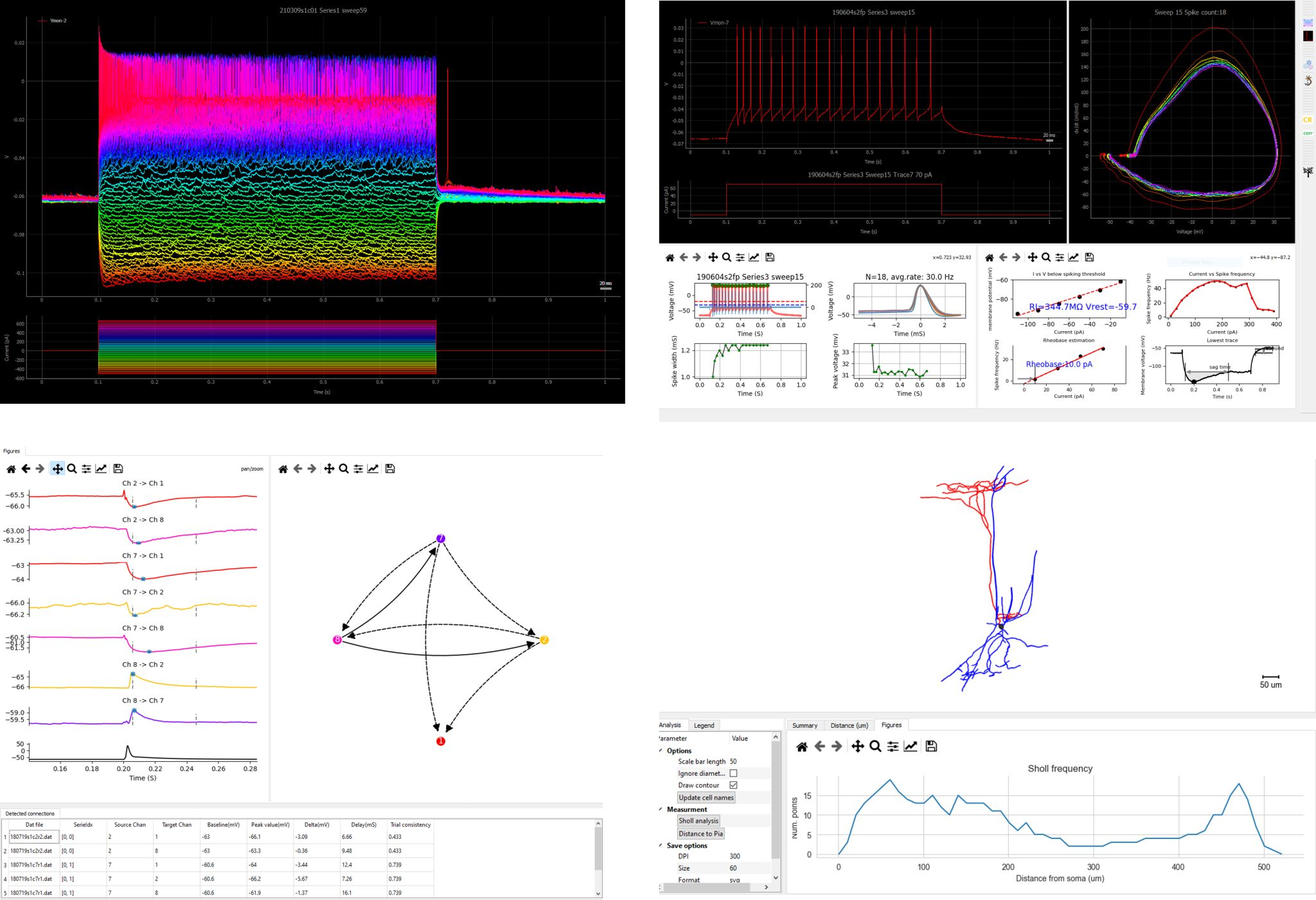
PatchView perform data analysis and visualization on multi channel whole-cell recording (multi-patch) data, including firing pattern analysis, event analysis, synaptic connection detection, morphological analysis and more.
- Free software: BSD 3-Clause license
- Documentation: https://patchview.readthedocs.io.
PatchView integrates multiple open-source tools (see credit page) and wrap them using an intuitive graphic user interface (GUI). Thus users can perform most analysis quickly for the data collected in a typical patch-clamp experiment without installing Python and these tools or writing any Python scripts.
- Importing both Heka data and Axon Instruments data. Exporting to Python pickle file or NWB (Neurodata Without Borders) file format.
- Visualizing single and multiple traces with zoom, pan operations.
- Automatically sorting experiments data according to predefined labels.
- Performing analysis on intrinsic membrane properties, action potential detection, firing pattern analysis.
- Synaptic connection analysis.
- Visualizing and quantification of neuron's morphological reconstruction from Neurolucida
If you find our work useful for your research, please cite:
Hu et al., (2022). PatchView: A Python Package for Patch-clamp Data Analysis and Visualization. Journal of Open Source Software, 7(78), 4706, https://doi.org/10.21105/joss.04706