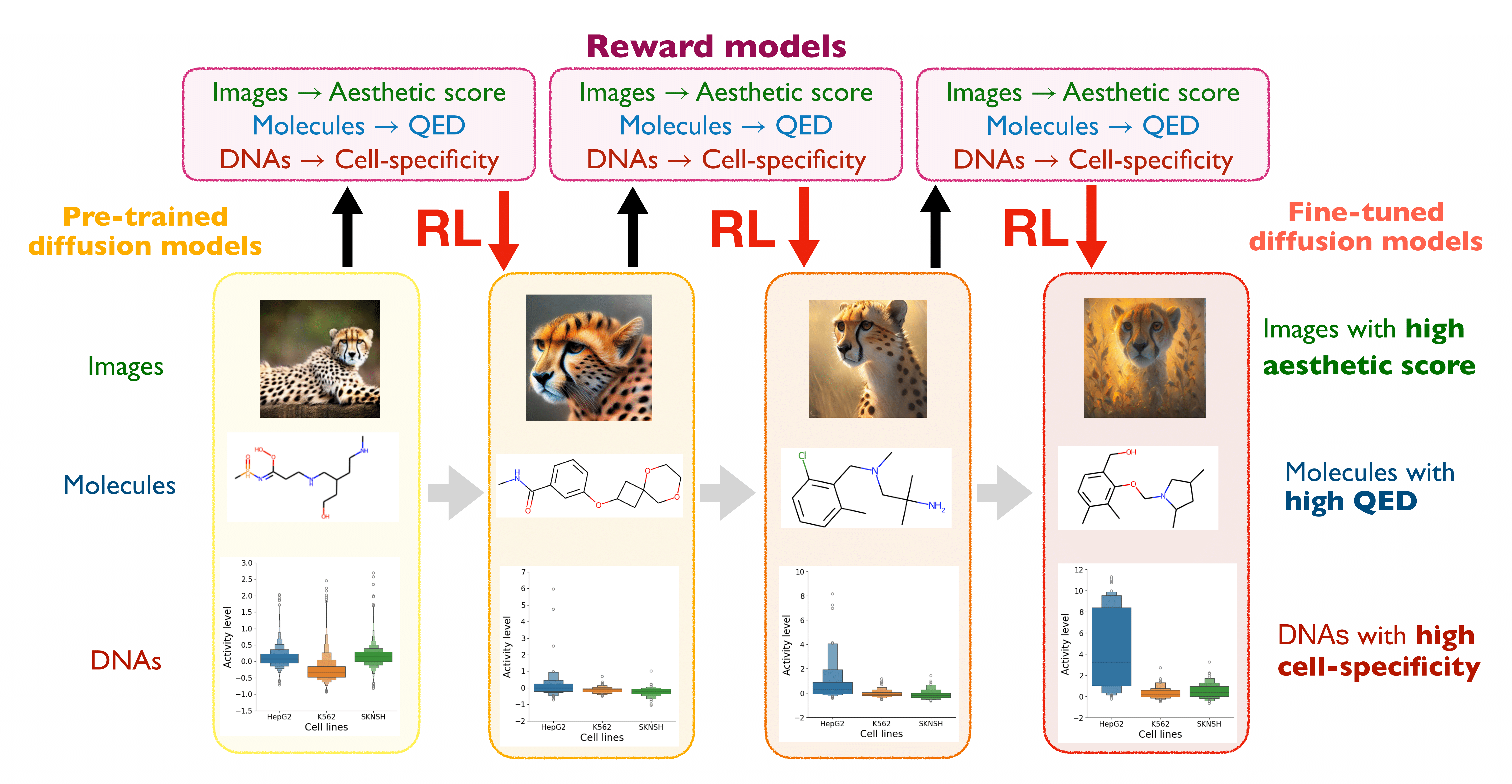
This code accompanies the tutorial/review paper on RL-based fine-tuning, where the objective is to maximize downstream reward functions by fine-tuning diffusion models with reinforcement learning (RL). In this implementation, we focus on the design of biological sequences, such as DNA (enhancers) and RNA (UTRs).
See ./tutorials. Each notebook is self-contained.
- 1-UTR_data.ipynb: Obtain raw data and proprocess.
- 2-UTR_diffusion_training.ipynb: Train conditional and unconditional diffuison models (Score-based Diffusion over Simplex).
- 3-UTR_evaluation.ipynb: Evaluate the performances of the pre-trained diffusion models.
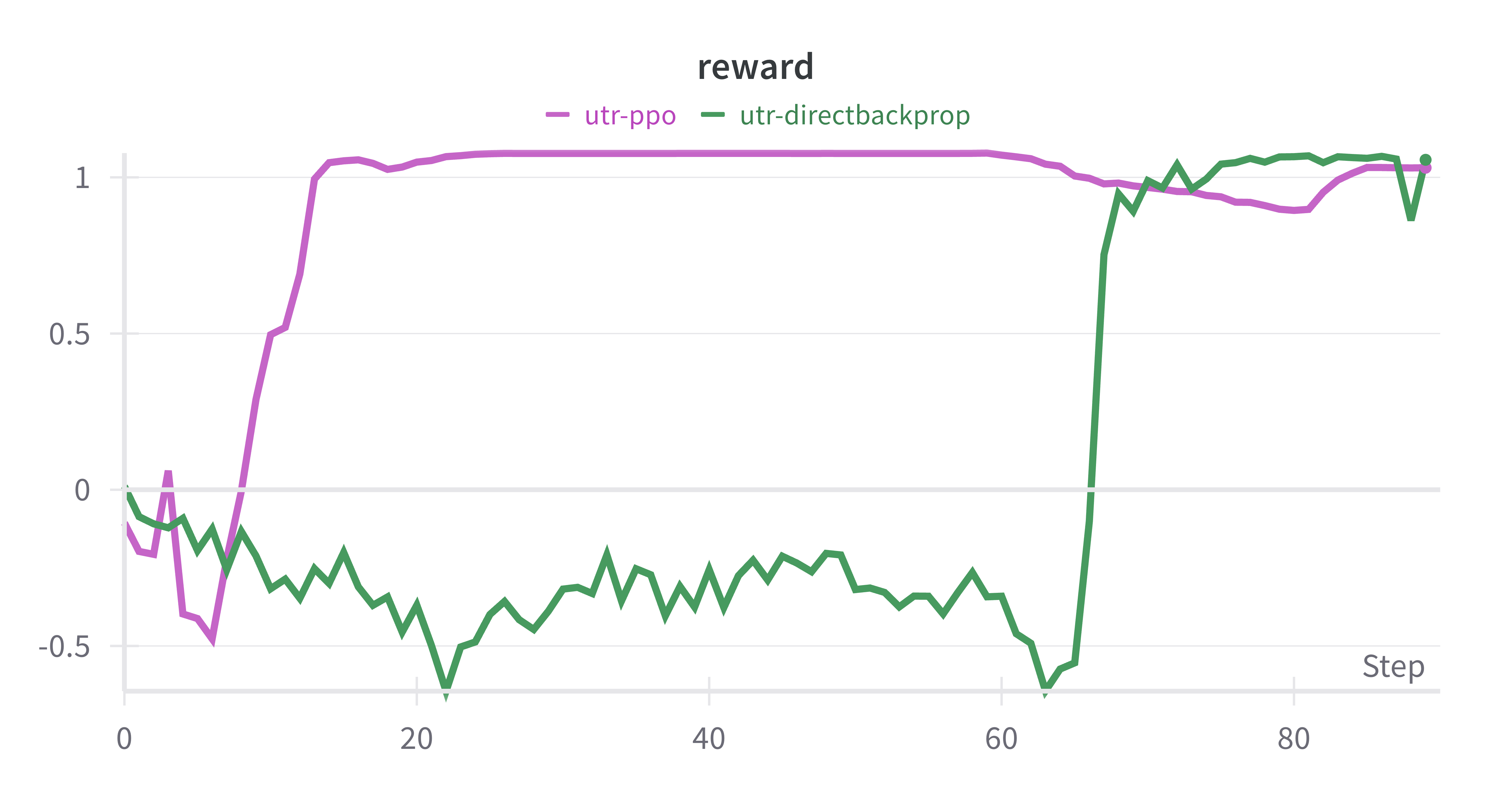
- 4-UTR_finetune_directbackprop.ipynb: Main fine-tuning code with direct reward backpropagation.
- 5-UTR_finetune_PPO.ipynb: Main fine-tuning code with PPO.
- Oracle_training: Train reward models from offline dataset, in which the samples are (x=sequence, y=activity level) pairs.
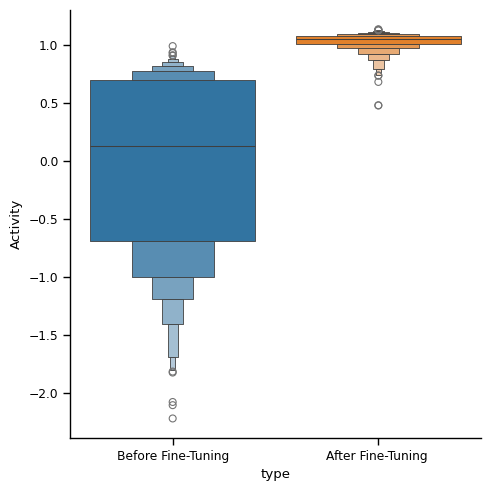
The following compares the generated RNA sequences (UTR )before/after fine-tuning. We optimize towards MRL (activity level).
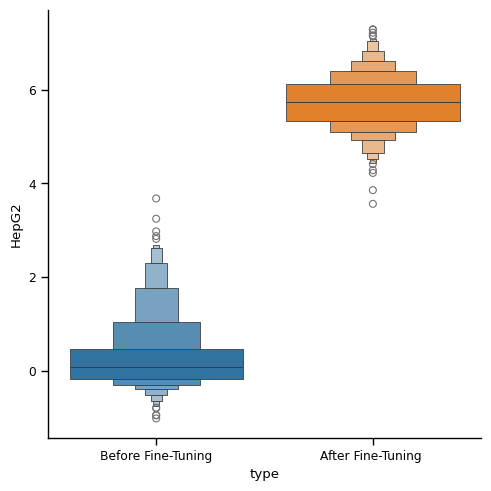
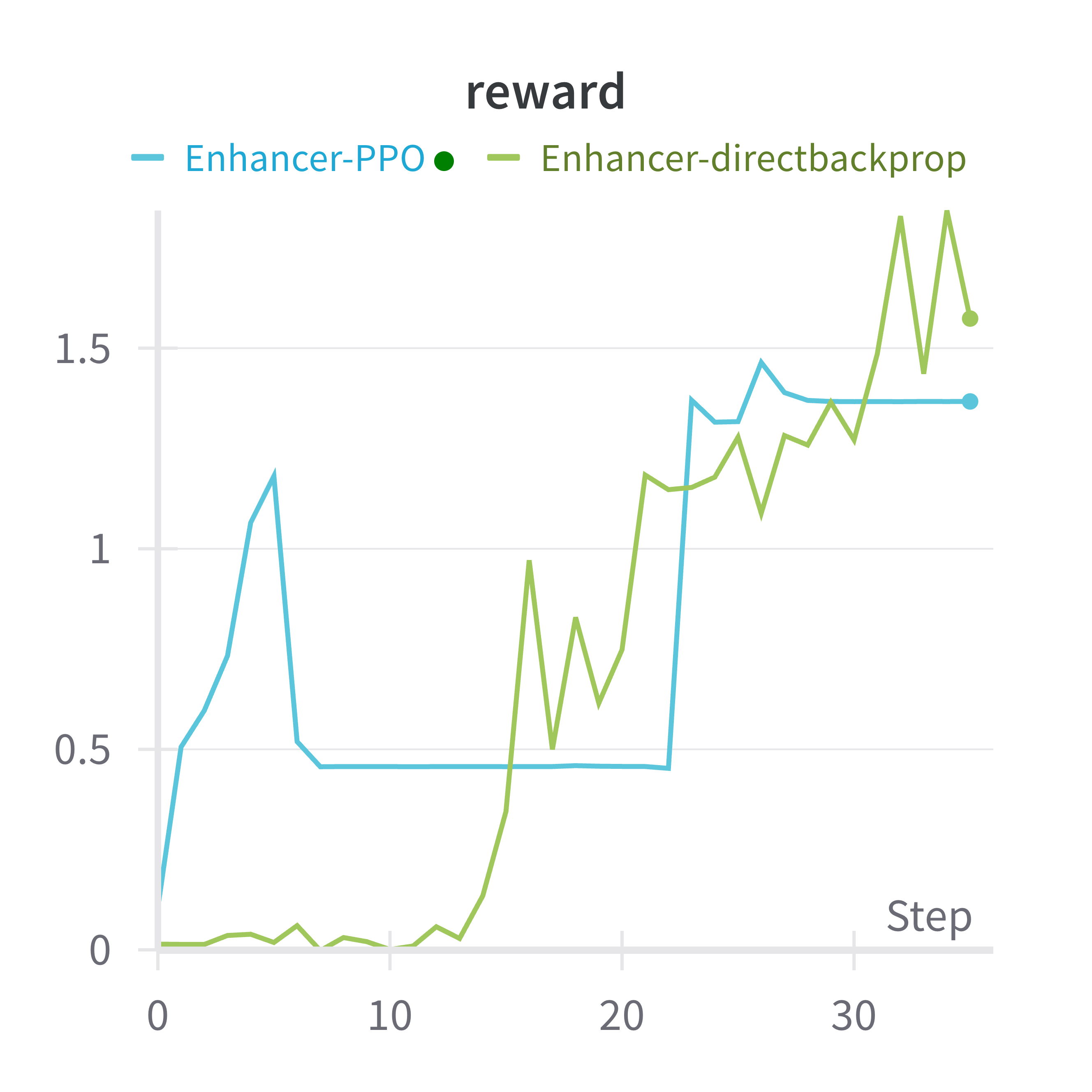
The following compares the generated DNA sequences (enhancer) before/after fine-tuning. We optimize towards activity level in HepG2 cell line.
-
We use Dirichlet Diffusion Models (Avedeyev et.al, 2023) as the backbone diffusion models. We acknowledge that our implementation is partly based on their codebase. We will implement more backbone diffusion models that are tailored to sequences.
-
Is over-optimization happening in the fine-tuning process?: Check paper BRAID (Uehera and Zhao et.al, 2024) on how to avoid it.
-
Lab-in-the-loop setting? : Check out SEIKO, implementation of online diffusion model fine-tuning, as well as the paper (Uehara and Zhao et.al, 2024).
- In training reward models, we use Enformers (Avset et al., 2021), one of the most common architectures for DNA sequence modeling. We use gReLU package for ease of implementation.
- The enhancer dataset is provided by Gosai et al., 2023.
- The UTR dataset is provided by Sample et al., 2019.
Create a conda environment with the following command:
conda create -n bioseq python = 3.12.0
conda activate bioseq
pip install -r requirements.txtIf you find this work useful in your research, please cite:
@article{Uehara2024understanding,
title={Understanding Reinforcement Learning-Based Fine-Tuning of Diffusion Models: A Tutorial and Review},
author={Uehara, Masatoshi and Zhao, Yulai and Biancalani, Tommaso and Levine, Sergey},
journal={arXiv preprint arXiv:2407.13734},
year={2024}
}