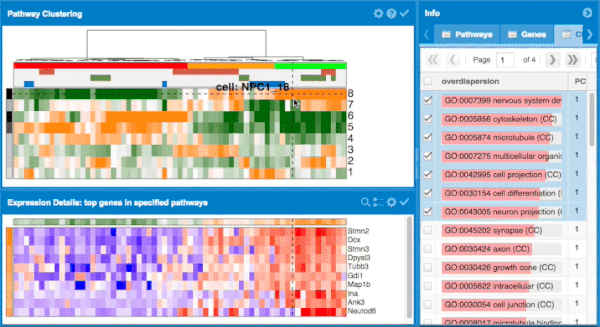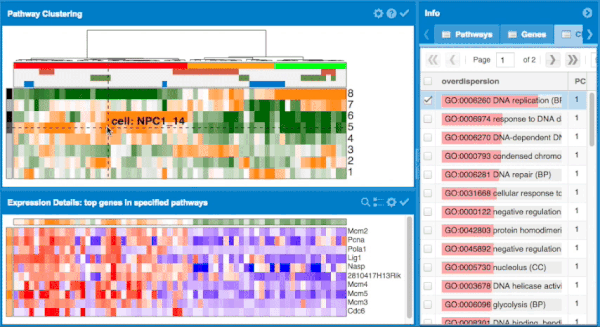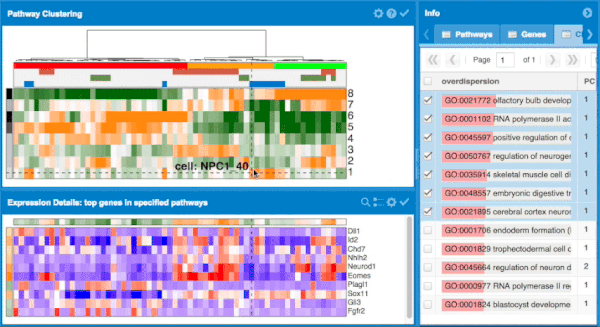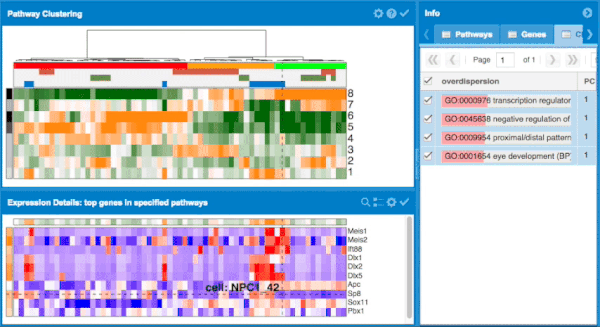
The scde package implements a set of statistical methods for analyzing single-cell RNA-seq data. scde fits individual error models for single-cell RNA-seq measurements. These models can then be used for assessment of differential expression between groups of cells, as well as other types of analysis. The scde package also contains the pagoda framework which applies pathway and gene set overdispersion analysis to identify aspects of transcriptional heterogeneity among single cells.
The overall approach to the differential expression analysis is detailed in the following publication:
"Bayesian approach to single-cell differential expression analysis" (Kharchenko PV, Silberstein L, Scadden DT, Nature Methods, doi:10.1038/nmeth.2967)
The overall approach to pathways and gene set overdispersion analysis is detailed in the following publication: "Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis" (Fan J, Salathia N, Liu R, Kaeser G, Yung Y, Herman J, Kaper F, Fan JB, Zhang K, Chun J, and Kharchenko PV, Nature Methods, doi:10.1038/nmeth.3734)
For additional installation information, tutorials, and more, please visit the SCDE website ☞
|
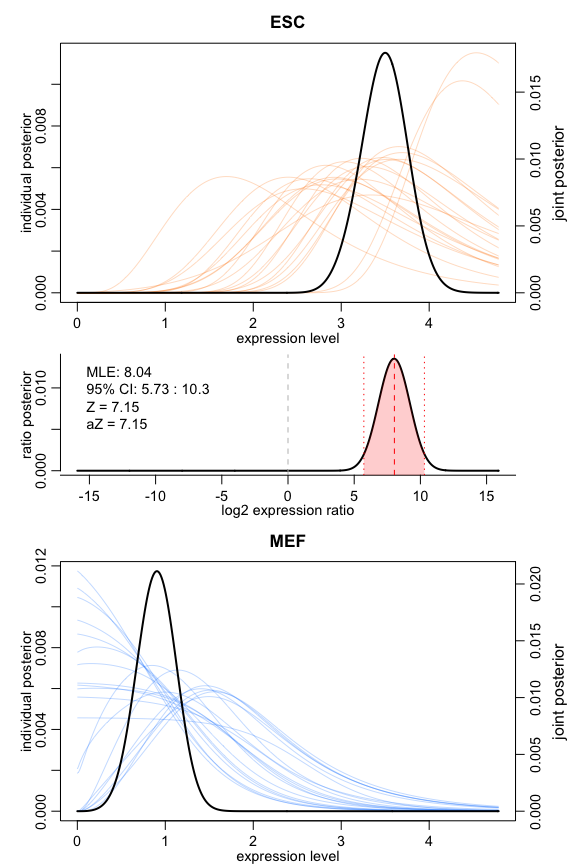
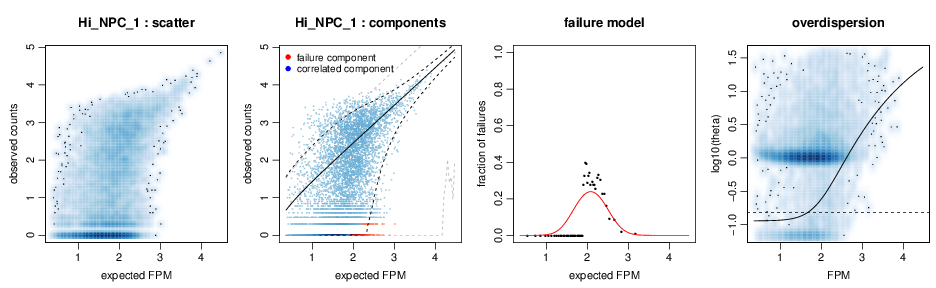
scde fits individual error models for single cells using counts derived from single-cell RNA-seq data to estimate drop-out and amplification biases on gene expression magnitude.
|
scde is maintained by Jean Fan of the Kharchenko Lab at the Department of Biomedical Informatics at Harvard Medical School.
We welcome any bug reports, enhancement requests, and other contributions. To submit a bug report or enhancement request, please use the scde GitHub issues tracker. For more substantial contributions, please fork this repo, push your changes to your fork, and submit a pull request with a good commit message. For more general discussions or troubleshooting, please consult the scde Google Group.