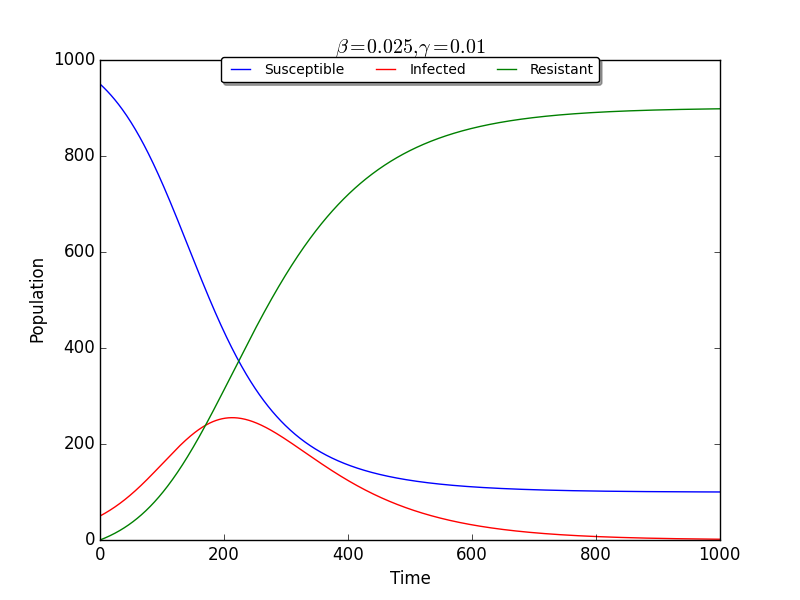
This project contains a basic SIR model, with build in plotting. A companion blog post for this project can be found here.
>>> import model
>>> m = model.SIR()
>>> m.run()
>>> m.plot()Specific parameters must be indicated when creating an new instance of the model.SIR class. For example, the beta (S to I) rate can be changed as follows:
>>> import model
>>> m = model.SIR(rateSI=0.05)Changable parameters include:
- 'eons' (number of time points to model, default 1000)
- 'Susceptible' (number of susceptible individuals at time 0, default 950)
- 'Infected' (number of infected individuals at time 0, default 50)
- 'Resistant' (number of resistant individuals at time 0, default 0)
- 'rateSI' (base rate 'beta' from S to I, default 0.05)
- 'rateIR' (base rate 'gamma' from I to R, default 0.01)
More complex alterations of the model will require specific engineering of the code. Feel free to dive in.