pgqc: A toolkit for quality control and adjustment of nucleotide redundancy in bacterial pan-genome estimates.
Pan-Genome quality control (PQQC) is a toolkit for evaluation of DNA sequence redundancy in analyzed pan-genomes (Panaroo or Roary).
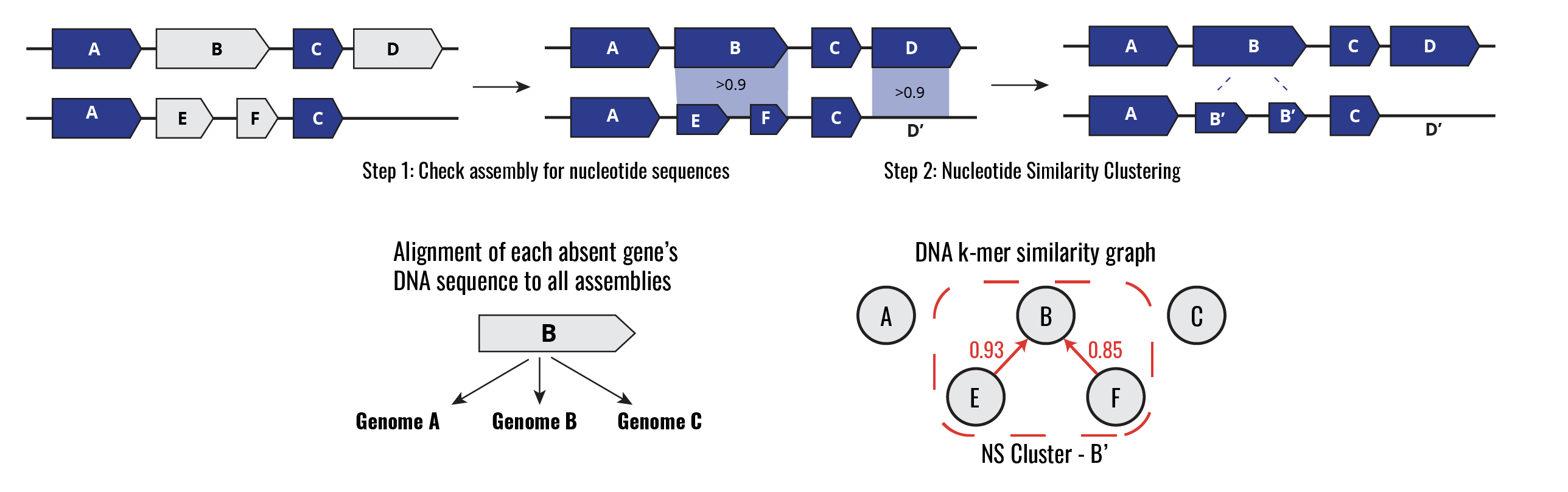
The PGQC Nucleotide Redundancy Analysis (PGQC-NRA) approach adjusts for redundancy at the DNA level in two steps (Methods). In step one, all genes predicted to be absent at the Amino Acid (AA) level are compared to their corresponding assembly at the nucleotide level. In cases where the nucleotide sequence is found with high coverage and sequence identity (Query Coverage & Sequence Identity > 90%), the gene is marked as “present at the DNA level”. Next, all genes are clustered and merged using a k-mer based metric of nucleotide similarity. Cases where two or more genes are divergent at the AA level but highly similar at the nucleotide level will be merged into a single “nucleotide similarity gene cluster”. After applying this method the pan-genome gene presence matrix is readjusted according to these results.
Currently, PGQC can be installed by cloning this repository and installing with pip.
git clone git@github.com:maxgmarin/pgqc.git
cd pgqc
pip install .
pgqc has 3 sub-commands: asmseqcheck, ava, nscluster
a) asmseqcheck - Perform alignment of all absent genes to all assemblies used in a pan-genome analysis.
usage: pgqc asmseqcheck [-h] -a IN_ASSEMBLIES -r IN_PG_REF -m IN_GENE_MATRIX -o OUT_GENE_MATRIX_WI_GENESEQCHECK [-c MIN_QUERY_COV] [-i MIN_SEQ_ID]
optional arguments:
-h, --help show this help message and exit
-a, --in_assemblies IN_ASSEMBLIES
Paths to input assemblies. (TSV)
-r, --in_pg_ref IN_PG_REF
Input pan-genome nucleotide reference. Typically output as `pan_genome_reference.fasta` by Panaroo/Roary (FASTA)
-m, --in_gene_matrix IN_GENE_MATRIX
Input pan-genome gene presence/absence matrix. Typically output as `gene_presence_absence.csv` by Panaroo/Roary (CSV)
-o, --out_gene_matrix_wi_geneseqcheck OUT_GENE_MATRIX_WI_GENESEQCHECK
Output pan-genome gene presence/absence matrix with updated gene presence/absence calls. (CSV). NOTE: 2 reflects that similar gene sequence is present at the nucleotide level (CSV)
-c, --min_query_cov MIN_QUERY_COV
Minimum query coverage to classify a gene as present within an assembly (0-1)
-i, --min_seq_id MIN_SEQ_ID
Minimum sequence identity to classify a gene as present within an assembly (0-1)
usage: pgqc ava [-h] -i IN_PG_REF -o OUT_AVA_TSV [-k KMER_SIZE]
optional arguments:
-h, --help show this help message and exit
-i, --in_pg_ref IN_PG_REF
Input Panaroo pan-genome nucleotide reference. Typically output as `pan_genome_reference.fasta` by Panaroo (FASTA)
-o, --out_ava_tsv OUT_AVA_TSV
All vs all comparison of sequence k-mer profiles. (TSV)
-k, --kmer_size KMER_SIZE
k-mer size (bp) to use for generating profile of unique k-mers for each sequence (Default: 31))
usage: pgqc nscluster [-h] -a IN_AVA_TSV -m IN_GENE_MATRIX_TSV [--min_ksim MIN_KSIM] -o OUT_NSC_GENE_MATRIX -c OUT_CLUSTERINFO_TSV
optional arguments:
-h, --help show this help message and exit
-a, --in_ava_tsv IN_AVA_TSV
Input table with all vs all comparison of sequence k-mer profiles. (TSV)
-m, --in_gene_matrix_tsv IN_GENE_MATRIX_TSV
Input pan-genome gene presence/absence matrix (CSV). NOTE: 2 reflects that similar gene sequence is present at the nucleotide level (CSV)
--min_ksim MIN_KSIM Minimum k-mer similarity (maximum Jaccard Similarity of k-mers between pair of sequences) to cluster sequences into the same "nucleotide similarity cluster" (Default: 0.8))
-o, --out_nsc_gene_matrix OUT_NSC_GENE_MATRIX
Nucleotide Similarity Cluster adjusted Gene Presence Matrix (TSV/Rtab)
-c, --out_clusterinfo_tsv OUT_CLUSTERINFO_TSV
Summary table with all genes that belong to a NSC (Nucleotide Similarity Cluster) (TSV)
Documentation is in progress.