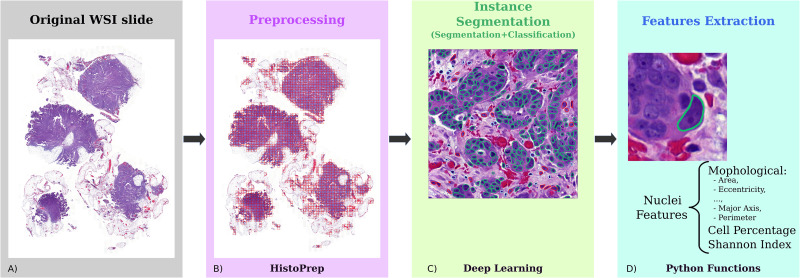
HEIP (HE-image-analysis-pipeline): set of tools to run end-to-end HE-image analysis
HEIP is a fully fledged pipeline for extracting cell level information from hematoxylin and eosin (H&E) whole slides. It consists of different steps and related tools ranging from HE-image patching, pre-processing, cell segmentation and classification, until downstream feature extraction. The pipeline, written entirely in Python, is modular and it is easy to add additional or modify existing steps.
To demonstrate the potential of HEIP, the pipeline was applied to analyze high-grade serous carcinoma H&E slides and the complete study is available in Journal of Pathology Informatics at this link
- Clone the repository
git clone git@github.com:ValeAri/HEIP_HE-image-analysis-pipeline.git-
cd to the repository
cd <path>/HEIP_HE-image-analysis-pipeline/ -
Create environment with python >3.7 and <3.11 (optional but recommended)
conda create --name HEIP python
conda activate HEIP
or
python3 -m venv HEIP
source HEIP/bin/activate
pip install -U pip
- Install dependencies
pip install -r requirements.txt
- Model
Instance segmenatation model available here!
- Patching HE images.
- Pre-process HE images.
- Train a segmentation model for HE images with a training set.
- Run inference with the segmentation model.
- Merge cell annotations.
- Morphological features extraction from all cells in the WSI.
NOTE You might want to modify the batchscripts to your needs (setting up right paths etc).
Training
cd HEIP_HE-image-analysis-pipeline/
sbatch ./batchscripts/train.shInference
cd HEIP_HE-image-analysis-pipeline/
sbatch ./batchscripts/infer_wsi.sh