For a given quality annotated read, the minEER algorithm identifies the subsequence which minimizes the expected error rate (EER; the mean q-score) while maximizing the subsequence length according to user defined EER and minimum sequence length thresholds. This procedure mimics the manual exercise of choosing truncation positions by glancing at a quality profile distribution, but ensures consistent results and is fast (can run on 20,000 files in under 15s). The minEER algorithm offers an improvement to current heuristic methods (sliding windows, quality score drop offs, etc.) which can be sensitive to noise and miss the opportunity for a deterministic solution based on reasonable criteria. The algorithm itself can be seen here.
The minEER pipeline (see documentation with mineer -h after installing) operates on an entire set of files from a single project. Assuming that all reads from a given direction (forward or reverse for paired reads) share a "similar" quality profile, the minEER pipeline runs the algorithm on a subsample of reads and to determine global truncation positions (where to start and end each read). All reads are then truncated according to these global positions and reads that fail to meet the user defined EER and minimum sequence lenght thresholds.
Two critical parameters to the minEER pipeline are the minimum acceptable length (MAL, set with flag --mal, default=100) and the maximum acceptable error (MAE, set with flag --mae, default=.01). See the Algorithm section below for an example of how these parameters operate.
Install with pip install mineer and then run mineer -h to view the input documentation and to test that installation worked properly.
After installing mineer, run the following:
# Download some fastq files to `sample_files/`
mineer-test-files
# Run the pipeline with default parameters (minimal acceptable error=.01)
mineer -i sample_files -f _1.fastq -r _2.fastq -o test_out -v sample_figs
Once you run the pipeline, a report of each step will appear as they execute. Files containing truncated reads will appear in the directory specified with -o. Providing the -v flag will produce visualizations like the following of quality profiles of untrimmed reads and the distribution of truncation positions identified by minEER:
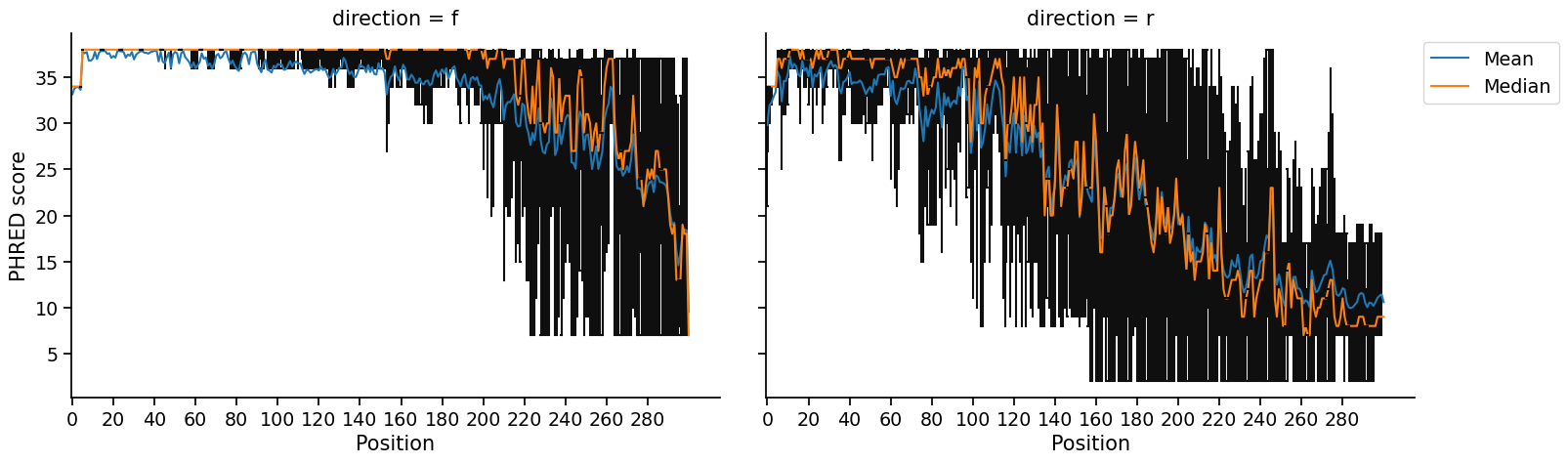
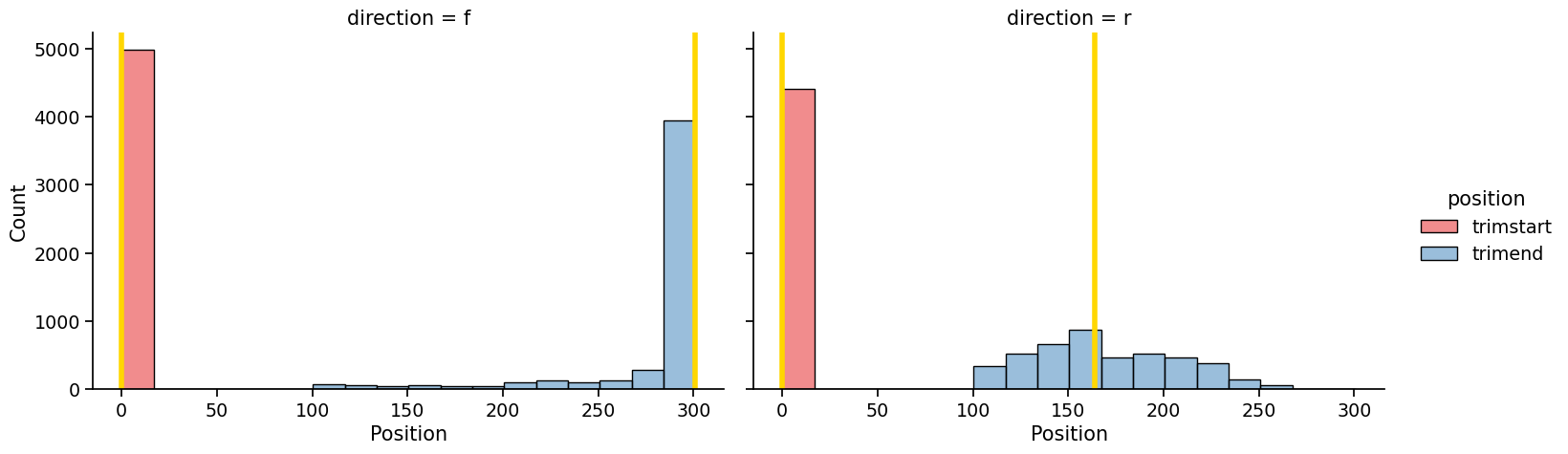
If you just want to compute truncation positions without writing out truncated files (which can take a while), then you can run mineer without writing files by not providing an output directory (no - o argument):
mineer -i sample_files -f _1.fastq -r _2.fastq
To produce visualizations without writing out truncated files, you can provide a directory with -v (again, without -o):
mineer -i sample_files -f _1.fastq -r _2.fastq -v sample_figs
Note that these examples use default parameters, which can be inspected with mineer -h.
Method:
- Ingest files and recognize pairs based on file names (using
xopen) - Run minEER on subset of reads
- Determine global truncation positions
- Truncate all reads to global positions and filter out read pairs that don't pass QC (currently requires longest length)
- Save truncated sequences
- Produce visualizations, if visualization output directory provided
For a given read, minEER works by finding the longest subsequence (with length => MAL) with an expected error rate <= MAE. The figure below illustrates the search space minEER explores to find the optimal subsequence with MAL=100 and MAE=1e-3. The expected error (EER) is calculated for each subsequence (at each start and end position) for all subsequences with length >= MAL (notice that all end positions are >= MAL) and then the longest subsequences with an expected error rate <= MAE is chosen (C shows all subsequence positions with expected error rate <= MAE in yellow).

Run tests with python -m unittest or pytest
Here is a longer list of SRRs to test on:
SRR9660346
SRR9660368
SRR9660375
SRR9660380
SRR9660372
SRR9660321
SRR9660322
SRR9660307
SRR9660387
SRR9660385