This project contains the source code and data for the paper titled "Dense reinforcement learning for safety validation of autonomous vehicles".
Dense-Deep-Reinforcement-Learning/
|__ conf: experiment configurations
|__ maps: maps for SUMO simulator
|__ checkpoints: model checkpoints for D2RL
|__ source_data: source data for constructing NDE and D2RL-based testing
|__ mtlsp: simulation platform
|__ envs: NDE and D2RL-based testing environments
|__ controller: vehicle controllers (e.g. IDM)
|__ data_analysis: refer to "Usage" section for detailed information
|__ main.py: main function for running NDE and D2RL-based testing
|__ utils.py: utility functions
|__ nadeinfoextractor.py: information extractor for logging experiment information
|__ requirements.txt: required packages
Feng, S., Sun, H., Yan, X., Zhu, H., Zou, Z., Shen, S., and Liu H.X. (2023). Dense reinforcement learning for safety validation of autonomous vehicles. Nature 615, 620–627. https://doi.org/10.1038/s41586-023-05732-2
@article{Feng2023,
title = {Dense reinforcement learning for safety validation of autonomous vehicles},
volume = {615},
url = {https://www.nature.com/articles/s41586-023-05732-2},
doi = {https://doi.org/10.1038/s41586-023-05732-2},
number = {7953},
journal = {Nature},
author = {Shuo Feng, Haowei Sun, Xintao Yan, Haojie Zhu, Zhengxia Zou, Shengyin Shen, Henry X. Liu},
year = {2023},
note = {Publisher: Nature Publishing Group},
pages = {620-627},
}
- Python installation
- This repository is developed and tested under python 3.10.4 on Ubuntu 20.04 system.
- Download all required datasets
- The user should download the
data_analysisfolder from here. Then, the user should merge the downloadeddata_analysisfolder with the originaldata_analysisfolder in the repo.
- The user should download the
git clone https://github.com/michigan-traffic-lab/Dense-Deep-Reinforcement-Learning.gitTo ensure high flexibility, it is recommended to use a virtual environment when running this repository. To set up the virtual environment, please follow the commands provided below:
virtualenv venv
source venv/bin/activateDue to the compatibility issue betwen gym and python setuptools, user should run follow commands to install a specific version of setuptools first:
pip install setuptools==65.5.0To install the Python packages required for this repository, execute the command provided below:
pip install -r requirements.txtIn order to use Jupyter notebooks for data analysis, it is necessary to have the ipykernel installed. To install it, users can execute the command provided below:
pip install ipykernelPlease refer to README_D2RL_Train.md for detailed information about D2RL training environment installation and detailed usage.
Please note that D2RL training process requries a different python environment compared to the python environment in README.md, with different python verison requirements and different python packages.
The project includes a data_analysis directory, which stores data (raw & processed), code (data processing and data analysis), and outcomes (figures) related to data analysis. The project analyzes various performance metrics such as time-to-collision, post-encroachment-time, bumper-to-bumper distance, crash rate, crash type, and crash severity. This section will focus on the post-encroachment-time (PET) analysis as an example, while the same procedure applies to all other performance metrics. Each performance metric analysis has a separate Jupyter notebook that contains its respective code.
For jupyter notebook usage, please refer to https://docs.jupyter.org/en/latest/
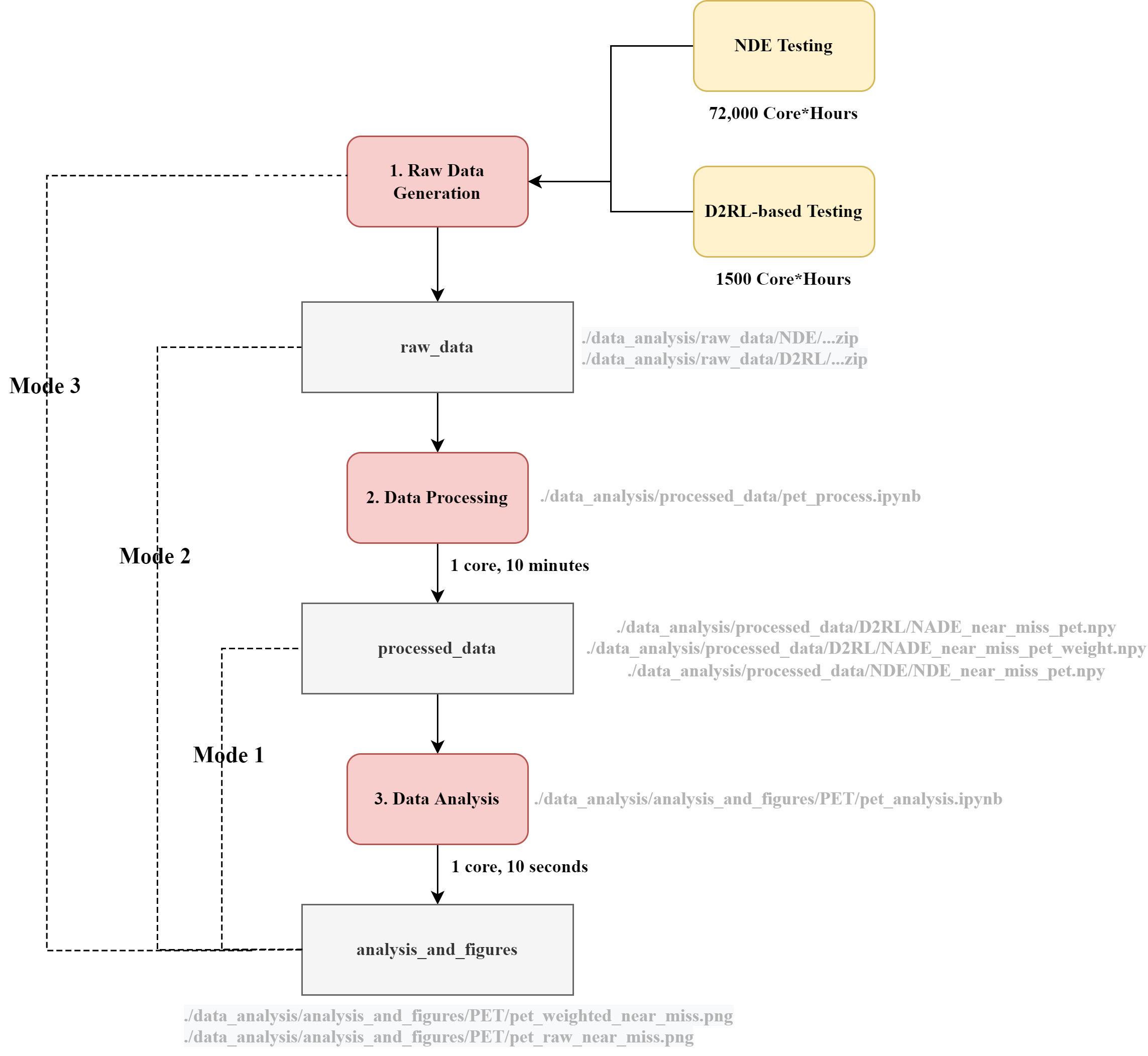
Since generating raw data is a time-consuming process (e.g., NDE testing experiment requires 72,000 core*hours), we have included the data generated during our experiments for users to quickly replicate the results. There are three running modes available:
- Mode 1 (recommended): data analysis using the data generated in our experiments;
- Mode 2: data processing and data analysis using the data generated in our experiments;
- Mode 3: raw data generation, data processing, and data analysis.
1 core*hour denotes the simulation running on one CPU core (Intel Xeon Gold 6154 3.0GHz) for one hour.
To provide further details of the three code running modes, a flowchart of PET data generation, processing, and analysis is provided as follows:
- For Mode 1: N/A
- For Mode 2: N/A
- For Mode 3:
- Please run the following commands to run the simulation and generate the raw experiment results for Naturalistic Driving Environment (NDE) testing and D2RL-based testing (the experiment_name can be specified by users):
-
python main.py --experiment_name 2lane_400m_NDE_testing --mode NDE # Use this for NDE Testing python main.py --experiment_name 2lane_400m_D2RL_testing --mode D2RL # Use this for D2RL Testing
- By default, the simulation result will be stored in
./data_analysis/raw_data/experiment_name.
-
- Please run the following commands to run the simulation and generate the raw experiment results for Naturalistic Driving Environment (NDE) testing and D2RL-based testing (the experiment_name can be specified by users):
-
For Mode 1: N/A
-
For Mode 2:
- Before running mode 2, the zipped dataset under
/data_analysis/raw_dataneeds to be unzipped to the same directory. Please note that the total size of the unzipped files will be around 130 GB. - Run all the code cells in the jupyter notebook (click "Run all" button in the menu bar of the notebook)
- The data processing code is stored in
/data_analysis/processed_data/. For example, the code for processing PET for both NDE experiments and D2RL experiments can be found in the jupyter notebookpet_process.ipynb, including several major steps:- Load raw experiment results
- Data processing: convert raw information (e.g., speed and position) to performance metrics (e.g., PET)
- Store the processed data in
/data_analysis/processed_data/NDEor/data_analysis/processed_data/D2RL - Newly generated files can be located after the completion of data processing:
NADE_near_miss_pet_weight.npyandNADE_near_miss_pet.npyunder/data_analysis/processed_data/D2RLNDE_near_miss_pet.npyunder/data_analysis/processed_data/NDE
- Before running mode 2, the zipped dataset under
-
For Mode 3:
- In the Jupyter notebook, please modify the following codes to analyse the recently generated experimental outcomes:
-
root_folder = "../raw_simulation_results/D2RL/" # Please change it to the position where you stored the newly generated raw experiment data path_list = ["Experiment-2lane_400m_IDM_NADE_2022-09-01"] # Please change it as the name of the folder generated in your new experiments
-
- After the modification, users can follow the data processing section of Mode 2:
- Run all the code cells in the jupyter notebook (click "Run all" button in the menu bar of the notebook)
- The data processing code is stored in
/data_analysis/processed_data/. For example, the code for processing PET for both NDE experiments and D2RL experiments can be found in the jupyter notebookpet_process.ipynb, including several major steps:- Load raw experiment results
- Data processing: convert raw information (e.g., speed and position) to performance metrics (e.g., PET)
- Store the processed data in
/data_analysis/processed_data/NDEor/data_analysis/processed_data/D2RL - Newly generated files can be located after the completion of data processing:
NADE_near_miss_pet_weight.npyandNADE_near_miss_pet.npyunder/data_analysis/processed_data/D2RLNDE_near_miss_pet.npyunder/data_analysis/processed_data/NDE
- In the Jupyter notebook, please modify the following codes to analyse the recently generated experimental outcomes:
This step is the same for all three running modes.
The directory /data_analysis/analysis_and_figures/ contains all the codes for data analysis and the figures generated. The file structure of the directory is shown below.
data_analysis/
|__ raw_data
|__ processed_data
|___analysis_and_figures
|______ crash_analysis
|_________ crash_severity_type_plot.ipynb # Analyze the crash severity and the crash type
|______ crash_rate
|_________ crash_rate_bootstrap_plot.ipynb # Analyze the crash rate, the convergency number
|______ near_miss_TTC_distance
|_________ ttc_distance_analysis_json.ipynb # Analyze the TTC, bumper-to-bumper distance
|______ PET
|_________ pet_analysis.ipynb # Analyze the PET
For example, the PET data analysis code can be found in /data_analysis/analysis_and_figures/PET/pet_analysis.ipynb, including following major steps:
- Load the processed experiment data from
/data_analysis/processed_data - Plot the PET histogram of D2RL experiments and NDE experiments, and then save figures to
/data_analysis/analysis_and_figures/PET.
Contributions are what make the open source community such an amazing place to be learn, inspire, and create. Any contributions you make are greatly appreciated.
- Fork the Project
- Create your Feature Branch (
git checkout -b feature/AmazingFeature) - Commit your Changes (
git commit -m 'Add some AmazingFeature') - Push to the Branch (
git push origin feature/AmazingFeature) - Open a Pull Request
This project is licensed under the [PolyForm Noncommercial License 1.0.0]. Please refer to LICENSE for more details.
H. L. and the team have filed a US provisional patent application 63/338,424.
Haowei Sun (haoweis@umich.edu)
Haojie Zhu (zhuhj@umich.edu)
Shuo Feng (fshuo@umich.edu)
For help or issues using the code, please create an issue for this repository or contact Haowei Sun (haoweis@umich.edu).
For general questions about the paper, please contact Henry Liu (henryliu@umich.edu).