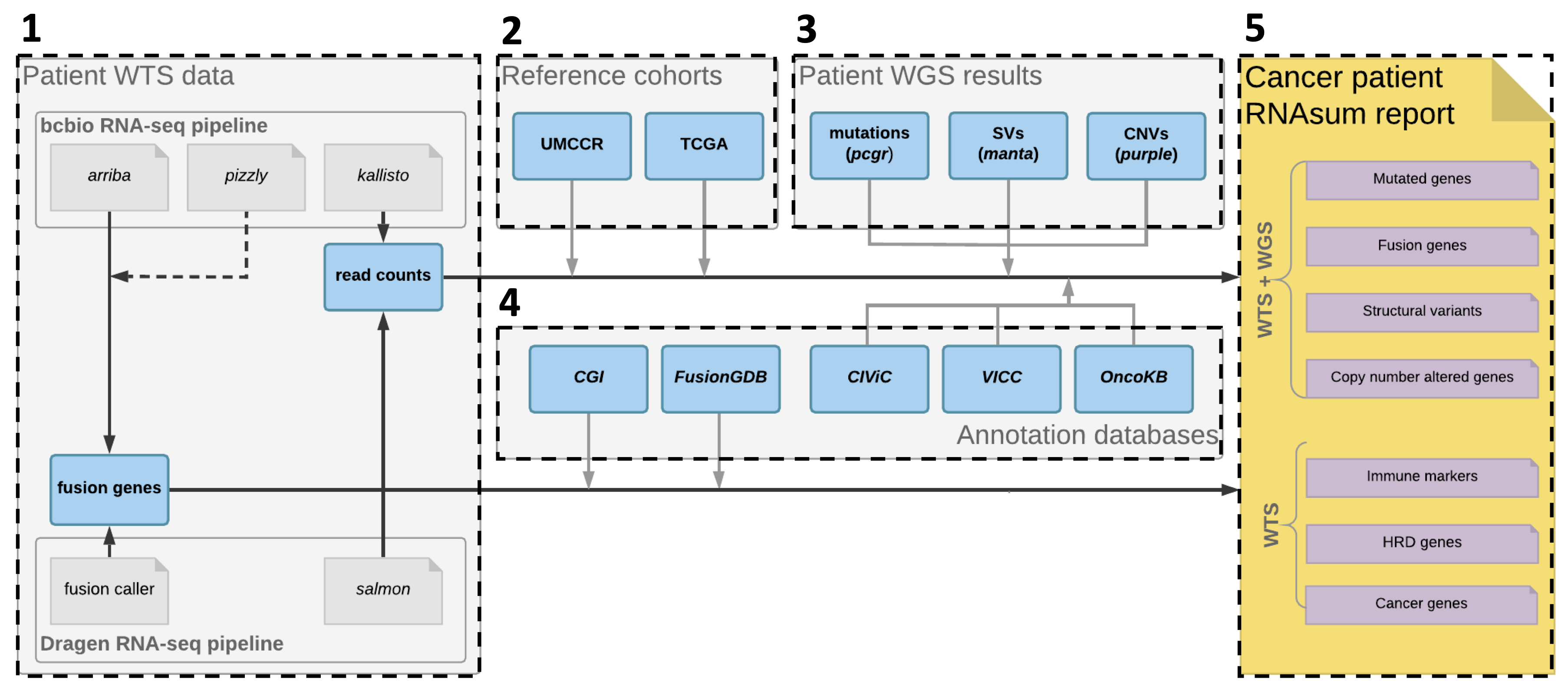
RNA-seq reporting workflow designed to post-process, summarise and visualise an output from bcbio-nextgen RNA-seq or Dragen RNA pipelines. Its main application is to complement genome-based findings from umccrise pipeline and to provide additional evidence for detected alterations.
Run the following to create a directory "rnasum" and install into it
mkdir rnasum
cd rnasum
source <(curl -s https://raw.githubusercontent.com/umccr/RNAseq-Analysis-Report/master/install.sh)
It will generate load_rnasum.sh script that can be sourced to load the rnasum environment:
source load_rnasum.sh
The pipeline consist of five main components illustrated and breifly described below. See the workflow.md for the complete description of the data processing workflow.
-
Collect patient sample WTS data from bcbio-nextgen RNA-seq or Dragen RNA pipeline including per-gene read counts and gene fusions.
-
Add expression data from reference cohorts to get an idea about expression levels of genes of interest in other cancer patient cohorts. The read counts are normalised, transformed and converted into a scale that allows to present the sample's expression measurements in the context of the reference cohorts.
-
Feed in genome-based findings from whole-genome sequencing (WGS) data to focus on genes of interest and provide additional evidence for dysregulation of mutated genes, or genes located within detected structural variants (SVs) or copy-number (CN) altered regions. The RNAsum pipeline is designed to be compatible with WGS patient report based on umccrise pipeline output.
-
Collate results with knowledge derived from in-house resources and public databases to provide additional source of evidence for clinical significance of altered genes, e.g. to flag variants with clinical significance or potential druggable targets.
-
The final product is a html-based interactive report with searchable tables and plots presenting expression levels of the genes of interest genes. The report consist of several sections described in report_structure.md.
The reference expression data is availbale for 33 cancer types and were derived from external (TCGA) and internal (UMCCR) resources.
In order to explore expression changes in queried sample we have built a high-quality pancreatic cancer reference cohort.
Depending on the tissue from which the patient's sample was taken, one of 33 cancer datasets from TCGA can be used as a reference cohort for comparing expression changes in genes of interest in investigated sample. Additionally, 10 samples from each of the 33 datasets were combined to create Pan-Cancer dataset, and for some cohorts extended sets are also available. All available datasets are listed in TCGA projects summary table. These datasets have been processed using methods described in TCGA-data-harmonization repository. The dataset of interest can be specified by using one of the TCGA project IDs (Project column) for the --dataset argument in RNAseq_report.R script (see Arguments section).
Each dataset was cleaned based on the quality metrics provided in the Merged Sample Quality Annotations file merged_sample_quality_annotations.tsv from TCGA PanCanAtlas initiative webpage (see TCGA-data-harmonization repository for more details, including sample inclusion criteria).
The publically available TCGA datasets are expected to demonstrate prominent batch effects when compared to the in-house WTS data due to differences in applied experimental procedures and analytical pipelines. Moreover, TCGA data may include samples from tissue material of lower quality and cellularity compared to samples processed using local protocols. To address these issues, we have built a high-quality internal reference cohort processed using the same pipelines as input data (see Data pre-processing section on the workflow page).
This internal reference set of 40 pancreatic cancer samples is based on WTS data generated at UMCCR and processed with bcbio-nextgen RNA-seq pipeline to minimise potential batch effects between investigated samples and the reference cohort and to make sure the data are comparable. The internal reference cohort assembly is summarised in Pancreatic-data-harmonization repository.
The are two rationales for using the internal reference cohort:
-
In case of pancreatic cancer samples this cohort is used (I) in batch effects correction, as well as (II) as a reference point for comparing per-gene expression levels observed in the investigated single-subject data and data from other pancreatic cancer patients.
-
In case of samples from any cancer type the data from the internal reference cohort is used in batch effects correction procedure performed to minimise technical-related variation in the data.
The pipeline accepts WTS data processed by bcbio-nextgen RNA-seq or Dragen RNA pipeline. Additionally, the WTS data can be integrated with WGS-based data processed using umccrise pipeline. In the latter case, the genome-based findings from corresponding sample are incorporated into the report and are used as a primary source for expression profiles prioritisation.
The only required WTS input data are read counts provided in quantification file from either bcbio-nextgen RNA-seq or Dragen RNA pipeline.
The read counts are provided within quantification file from kallisto (see example abundance.tsv file and its description). The per-transcript abundances are reported in estimated counts (est_counts) and in Transcripts Per Million (tpm), which are then converted to per-gene estimates. Additionally, a list of fusion genes detected by arriba and pizzly can be provided (see example fusions.tsv and test_sample_WTS-flat.tsv).
Table below lists all input data accepted in the pipeline:
| Input file | Tool | Example | Required |
|---|---|---|---|
| Quantified abundances of transcripts | kallisto | abundance.tsv | Yes |
| List of detected fusion genes | arriba pizzly |
fusions.tsv test_sample_WTS-flat.tsv |
No |
| Plots of detected fusion genes using arriba | arriba | fusions.pdf | No |
These files are expected to be organised following the folder structure below
|
|____<SampleName>
|____kallisto
| |____abundance.tsv
|____pizzly
| |____<SampleName>-flat.tsv
|____arriba
|____fusions.pdf
|____fusions.tsv
Fusion genes detected by pizzly are expected to be listed in the flat table. By default two output tables are provided: (1) <sample_name>-flat.tsv listing all gene fusion candidates and (2) <sample_name>-flat-filtered.tsv listing only gene fusions remaining after filtering step. However, this workflow makes use of gene fusions listed in the unfiltered pizzly output file (see example test_sample_WTS-flat.tsv) since it was noted that some genuine fusions (based on WGS data and curation efforts) are excluded in the filtered pizzly output file.
The read counts are provided within quantification file from salmon (see example TEST.quant.sf file and its description). The per-transcript abundances are reported in estimated counts (NumReads ) and in Transcripts Per Million (TPM ), which are then converted to per-gene estimates. Additionally, a list of fusion genes can be provided (see example TEST.fusion_candidates.final).
Table below lists all input data accepted in the pipeline:
| Input file | Tool | Example | Required |
|---|---|---|---|
| Quantified abundances of transcripts | salmon | TEST.quant.sf | Yes |
| List of detected fusion genes | Dragen RNA | TEST.fusion_candidates.final | No |
These files are expected to be organised following the folder structure below
|
|____<SampleName>
|____<SampleName>quant.sf
|____<SampleName>.fusion_candidates.final
The following umccrise output files are accepted as input data in the pipeline:
| Input file | Tool | Example | Required |
|---|---|---|---|
| List of detected and annotated single-nucleotide variants (SNVs) and indels | PCGR | test_subject__test_sample_WGS-somatic.pcgr.snvs_indels.tiers.tsv | No |
| List of genes involved in CN altered regions | PURPLE | test_subject__test_sample_WGS.purple.gene.cnv | No |
| List of genes involved in SV regions | Manta | test_subject__test_sample_WGS-sv-prioritize-manta-pass.tsv | No |
These files are expected to be organised following the folder structure below
|
|____umccrised
|____<SampleName>
|____pcgr
| |____<SampleName>-somatic.pcgr.snvs_indels.tiers.tsv
|____purple
| |____<SampleName>.purple.gene.cnv
|____structural
|____<SampleName>-manta.tsv
To run the pipeline execure the RNAseq_report.R script. This script catches the arguments from the command line and passes them to the RNAseq_report.Rmd script to produce the interactive HTML report.
| Argument | Description | Required |
|---|---|---|
| --sample_name | The name of the sample to be analysed and reported | Yes |
| --bcbio_rnaseq | Location of the results folder from bcbio-nextgen RNA-seq pipeline | Yes* |
| --dragen_rnaseq | Location of the results folder from Dragen RNA pipeline | Yes* |
| --report_dir | Desired location for the report | Yes |
| --dataset | Dataset to be used as external reference cohort. Available options are TCGA project IDs listed in TCGA projects summary table Project column (default is PANCAN) |
No |
| --transform | Transformation method for converting read counts. Available options are: CPM (default) and TPM |
No |
| --norm | Normalisation method. Available options are: TMM (default), TMMwzp, RLE, upperquartile or none for CPM-transformed data, and quantile (default) or none for TPM-transformed data |
No |
| --batch_rm | Remove batch-associated effects between datasets. Available options are: TRUE (default) and FALSE |
No |
| --filter | Filtering out low expressed genes. Available options are: TRUE (default) and FALSE |
No |
| --log | Log (base 2) transform data before normalisation. Available options are: TRUE (default) and FALSE |
No |
| --scaling | Apply gene-wise (default) or group-wise data scaling. Available options are: TRUE (default) and FALSE |
No |
| --drugs | Include drug matching section in the report. Available options are: TRUE and FALSE (default) |
No |
| --immunogram | Include immunogram in the report. Available options are: TRUE and FALSE (default) |
No |
| --umccrise | Location of the corresponding umccrise output (including PCGR (see example), Manta (see example) and PURPLE (see example) output files) from genome-based data | No |
| --pcgr_tier | Tier threshold for reporting variants reported in PCGR (if PCGR results are available, default is 4) |
No |
| --pcgr_splice_vars | Include non-coding splice_region_variants reported in PCGR (if PCGR results are available). Available options are: TRUE (default) and FALSE |
No |
| --cn_loss | CN threshold value to classify genes within lost regions (if CN results from PURPLE are available, default is 5th percentile of all CN values) |
No |
| --cn_gain | CN threshold value to classify genes within gained regions (if CN results from PURPLE are available, default is 95th percentile of all CN values) |
No |
| --clinical_info | Location of xslx file with clinical information (see example ) | No |
| --clinical_id | ID required to match sample with the subject clinical information (if available) | No |
| --subject_id | Subject ID. Note, if umccrise is specified (flag --umccrise) then subject ID is extracted from umccrise output files and used to overwrite this argument |
No |
| --sample_source | Source of investigated sample (e.g. fresh frozen tissue, organoid; for annotation purposes only) | No |
| --sample_name_mysql | Desired sample name for MySQL insert command. By default value in --sample_name is used |
No |
| --project | Project name (for annotation purposes only) | No |
| --top_genes | The number of top ranked genes to be presented (default is 5) |
No |
| --dataset_name_incl | Include dataset in the report name. Available options are: TRUE and FALSE (default) |
No |
| --save_tables | Save interactive summary tables as HTML files. Available options are: TRUE (default) and FALSE |
No |
| --hide_code_btn | Hide the Code button allowing to show/hide code chunks in the final HTML report. Available options are: TRUE (default) and FALSE |
No |
| --grch_version | Human reference genome version used for genes annotation. Available options: 37 and 38 (default) |
No |
* Location of the results folder from either bcbio-nextgen RNA-seq or Dragen RNA pipeline is required.
Packages: required packages are listed in environment.yaml file.
Human reference genome GRCh38 (Ensembl based annotation version 86) is used for genes annotation as default. Alternatively, human reference genome GRCh37 (Ensembl based annotation version 75) is used when argument grch_version is set to 37.
Below are command line use examples for generating Patient Transcriptome Summary report using:
- make sure that the created conda environment (see Installation section) is activated
conda activate rnasum
- RNAseq_report.R script (see the beginning of Usage section) should be executed from rmd_files folder
cd rmd_files
- example data is provided in data/test_data folder
- Usually the data processing and report generation would take less than 20 minutes using 16GB RAM memory and 1 CPU
In this scenario, only WTS data will be used and only expression levels of key Cancer genes, Fusion genes, Immune markers and homologous recombination deficiency genes (HRD genes) will be reported. Moreover, gene fusions reported in Fusion genes section will not contain inforamation about evidence from genome-based data. A subset of the TCGA pancreatic adenocarcinoma dataset is used as reference cohort (--dataset TEST ).
The input files are expected to be organised following the folder structure described in Input data:WTS section.
Rscript RNAseq_report.R --sample_name test_sample_WTS --dataset TEST --bcbio_rnaseq $(pwd)/../data/test_data/final/test_sample_WTS --report_dir $(pwd)/../data/test_data/final/test_sample_WTS/RNAsum --save_tables FALSE
The interactive HTML report named
test_sample_WTS.RNAsum.htmlwill be created indata/test_data/final/test_sample_WTS/RNAsumfolder.
Rscript RNAseq_report.R --sample_name test_sample_WTS --dataset TEST --dragen_rnaseq $(pwd)/../data/test_data/stratus/test_sample_WTS --report_dir $(pwd)/../data/test_data/stratus/test_sample_WTS/RNAsum --save_tables FALSE
The interactive HTML report named
test_sample_WTS.RNAsum.htmlwill be created indata/test_data/stratus/test_sample_WTS/RNAsumfolder.
This is the most frequent and preferred case, in which the WGS-based findings will be used as a primary source for expression profiles prioritisation. The genome-based results can be incorporated into the report by specifying location of the corresponding umccrise output files (including results from PCGR, PURPLE and Manta) using --umccrise argument. The Mutated genes, Structural variants and CN altered genes sections will contain information about expression levels of the mutated genes, genes located within detected structural variants (SVs) and copy-number (CN) altered regions, respectively. The results in the Fusion genes section will be ordered based on the evidence from genome-based data. A subset of the TCGA pancreatic adenocarcinoma dataset is used as reference cohort (--dataset TEST ).
The umccrise output files are expected to be organised following the folder structure described in Input data:WGS section.
Rscript RNAseq_report.R --sample_name test_sample_WTS --dataset TEST --bcbio_rnaseq $(pwd)/../data/test_data/final/test_sample_WTS --report_dir $(pwd)/../data/test_data/final/test_sample_WTS/RNAsum --umccrise $(pwd)/../data/test_data/umccrised/test_subject__test_sample_WGS --save_tables FALSE
The interactive HTML report named
test_sample_WTS.RNAsum.htmlwill be created indata/test_data/final/test_sample_WTS/RNAsumfolder.
Rscript RNAseq_report.R --sample_name test_sample_WTS --dataset TEST --dragen_rnaseq $(pwd)/../data/test_data/stratus/test_sample_WTS --report_dir $(pwd)/../data/test_data/stratus/test_sample_WTS/RNAsum --umccrise $(pwd)/../data/test_data/umccrised/test_subject__test_sample_WGS --save_tables FALSE
The interactive HTML report named
test_sample_WTS.RNAsum.htmlwill be created indata/test_data/stratus/test_sample_WTS/RNAsumfolder.
For samples derived from subjects, for which clinical information is available, a treatment regimen timeline can be added to the Patient Transcriptome Summary report. This can be added by specifying location of a relevant excel spreadsheet (see example test_clinical_data.xlsx) using the --clinical_info argument. In this spreadsheet, at least one of the following columns is expected: NEOADJUVANT REGIMEN, ADJUVANT REGIMEN, FIRST LINE REGIMEN, SECOND LINE REGIMEN or THIRD LINE REGIMEN, along with START and STOP dates of corresponding treatments. A subset of the TCGA pancreatic adenocarcinoma dataset is used as reference cohort (--dataset TEST ).
Rscript RNAseq_report.R --sample_name test_sample_WTS --dataset TEST --bcbio_rnaseq $(pwd)/../data/test_data/final/test_sample_WTS --report_dir $(pwd)/../data/test_data/final/test_sample_WTS/RNAsum --umccrise $(pwd)/../data/test_data/umccrised/test_subject__test_sample_WGS --clinical_info $(pwd)/../data/test_data/test_clinical_data.xlsx --save_tables FALSE
The interactive HTML report named
test_sample_WTS.RNAsum.htmlwill be created indata/test_data/final/test_sample_WTS/RNAsumfolder.
Rscript RNAseq_report.R --sample_name test_sample_WTS --dataset TEST --dragen_rnaseq $(pwd)/../data/test_data/stratus/test_sample_WTS --report_dir $(pwd)/../data/test_data/stratus/test_sample_WTS/RNAsum --umccrise $(pwd)/../data/test_data/umccrised/test_subject__test_sample_WGS --clinical_info $(pwd)/../data/test_data/test_clinical_data.xlsx --save_tables FALSE
The interactive HTML report named
test_sample_WTS.RNAsum.htmlwill be created indata/test_data/stratus/test_sample_WTS/RNAsumfolder.
The pipeline generates html-based Patient Transcriptome Summary report and results folder within user-defined output folder:
|
|____<output>
|____<SampleName>.<output>.html
|____results
|____exprTables
|____glanceExprPlots
|____...
The generated html-based Patient Transcriptome Summary report includes searchable tables and interactive plots presenting expression levels of altered genes, as well as links to public resources describing the genes of interest. The report consist of several sections, including:
- Input data
- Clinical information*
- Findings summary
- Mutated genes**
- Fusion genes
- Structural variants**
- CN altered genes**
- Immune markers
- HRD genes
- Cancer genes
- Drug matching
- Addendum
* if clinical information is available; see --clinical_info argument
** if genome-based results are available; see --umccrise argument
Detailed description of the report structure, including results prioritisation and visualisation is available in report_structure.md.
The results folder contains intermediate files, including plots and tables that are presented in the report.
- Pull ready to run docker image from DockerHub
docker pull umccr/rnasum:0.3.2
- An example command to use this pulled docker container is:
docker run --rm -v /path/to/RNAseq-report/RNAseq-Analysis-Report/envm/wts-report-wrapper.sh:/work/test.sh -v /path/to/RNAseq-report/RNAseq-Analysis-Report/data:/work c18db89d3093 /work/test.sh
-
Assumptions
- You are running the RNAsum container against the RNAsum code and test/reference data