Paper | OpenReview | Poster | Slides
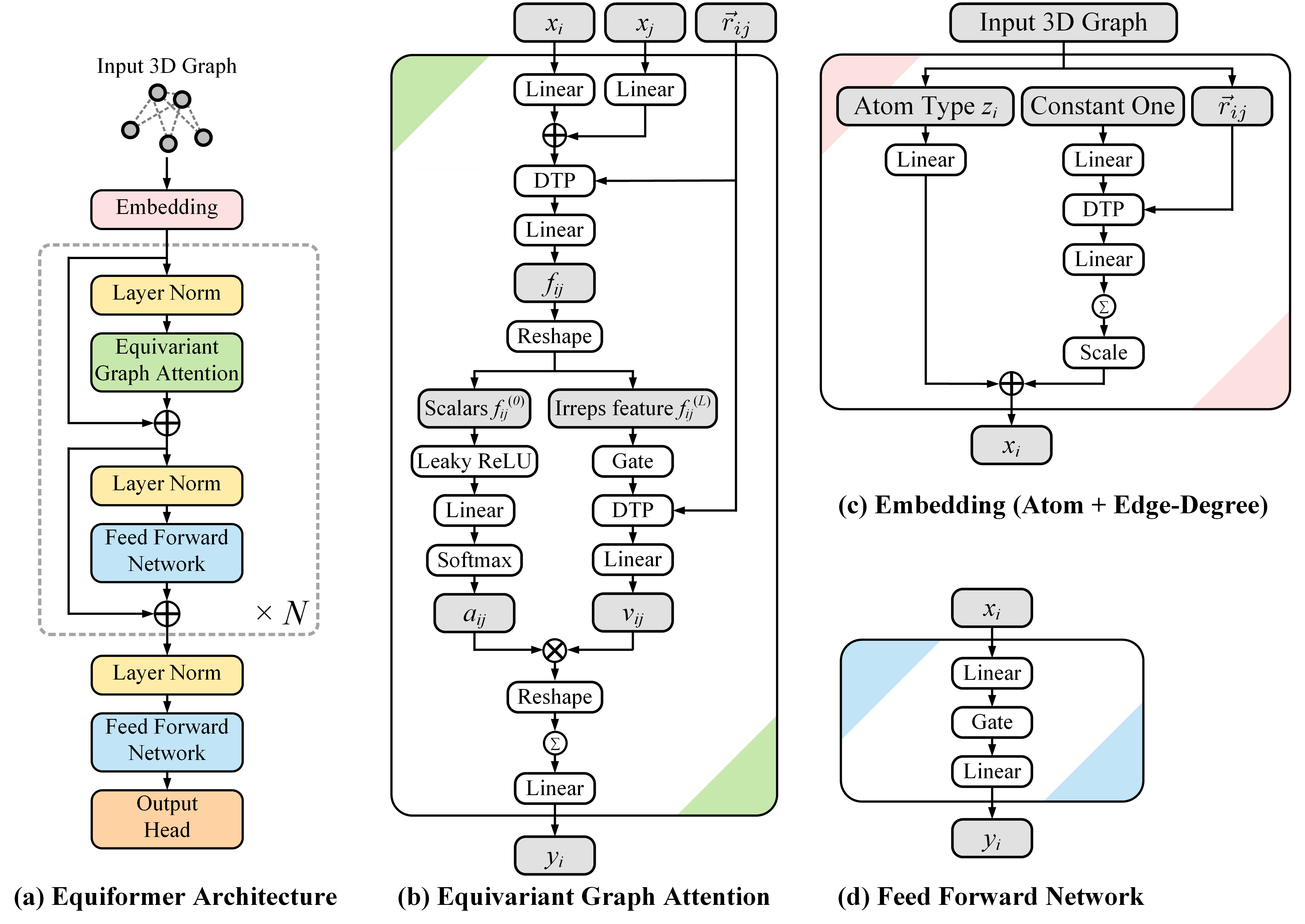
This repository contains the official PyTorch implementation of the work "Equiformer: Equivariant Graph Attention Transformer for 3D Atomistic Graphs" (ICLR 2023 Spotlight).
See here for setting up the environment.
The dataset of QM9 will be automatically downloaded when running training.
The dataset of MD17 will be automatically downloaded when running training.
The dataset for different tasks can be downloaded by following instructions in their GitHub repository.
After downloading, place the datasets under datasets/oc20/ by using ln -s.
Take is2re as an example:
cd datasets
mkdir oc20
cd oc20
ln -s ~/ocp/data/is2re is2re-
We provide training scripts under
scripts/train/qm9/equiformer. For example, we can train Equiformer for the task ofalphaby running:sh scripts/train/qm9/equiformer/target@1.sh
-
The QM9 dataset will be downloaded automatically as we run training for the first time.
-
The target number for different regression tasks can be found here.
-
We also provide the code for training Equiformer with linear messages and dot product attention. To train Equiformer with linear messages, replace
--model-name 'graph_attention_transformer_nonlinear_l2'with--model-name 'graph_attention_transformer_l2'in training scripts. -
The training scripts for Equiformer with linear messages and dot product attention can be found in
scripts/train/qm9/dp_equiformer. -
Training logs of Equiformer can be found here.
-
We provide training scripts under
scripts/train/md17/equiformer. For example, we can train Equiformer for the molecule ofaspirinby running:sh scripts/train/md17/equiformer/se_l2/target@aspirin.sh # L_max = 2 sh scripts/train/md17/equiformer/se_l3/target@aspirin.sh # L_max = 3
-
Training logs of Equiformer with
$L_{max} = 2$ and$L_{max} = 3$ can be found here ($L_{max} = 2$ ) and here ($L_{max} = 3$ ). Note that the units of energy and force are kcal mol$^{-1}$ and kcal mol$^{-1}$ Å$^{-1}$ and that we report energy and force in units of meV and meV Å$^{-1}$ in the paper.
-
We train Equiformer on IS2RE data only by running:
sh scripts/train/oc20/is2re/graph_attention_transformer/l1_256_nonlinear_split@all_g@2.sh
a. This requires 2 GPUs and results in energy MAE of around 0.5088 eV for the ID sub-split of the validation set.
b. Pretrained weights and training logs can be found here.
-
We train Equiformer on IS2RE data with IS2RS auxiliary task and Noisy Nodes data augmentation by running:
sh scripts/train/oc20/is2re/graph_attention_transformer/l1_256_blocks@18_nonlinear_aux_split@all_g@4.sh
a. This requires 4 GPUs and results in energy MAE of around 0.4156 eV for the ID sub-split of the validation set.
b. Pretrained weights and training logs can be found here.
We have different files and models for QM9, MD17 and OC20.
netsincludes code of different network architectures for QM9, MD17 and OC20.scriptsincludes scripts for training models for QM9, MD17 and OC20.
main_qm9.pyis the training code for QM9 dataset.
main_md17.pyis the code for training and evaluation on MD17 dataset.
Some differences are made to support:
- Removing weight decay for certain parameters specified by
no_weight_decay. One example is here. - Cosine learning rate.
main_oc20.pyis the code for training and evaluating.oc20/trainercontains the code for energy trainers.oc20/configscontains the config files for IS2RE.
Our implementation is based on PyTorch, PyG, e3nn, timm, ocp, SEGNN and TorchMD-NET.
If you use our code or method in your work, please consider citing the following:
@inproceedings{
liao2023equiformer,
title={Equiformer: Equivariant Graph Attention Transformer for 3D Atomistic Graphs},
author={Yi-Lun Liao and Tess Smidt},
booktitle={International Conference on Learning Representations},
year={2023},
url={https://openreview.net/forum?id=KwmPfARgOTD}
}Please direct any questions to Yi-Lun Liao (ylliao@mit.edu).