MAPpoly-MP is an under-development package to build genetic maps in diploids and polyploid interconnected populations. Some of the MAPpoly-MP features:
- Mutli-population
- Multi-allelic (from biallelic to complete informative markers)
- Multi-ploidy (2, 4, 6, and any combination of those)
The core C++ function to re-estimate a genetic map and compute the haplotype probabilities in any of the above situations is implemented. However, several functions must be implemented before releasing the package to the general public. In the next sections, I will present some functionality implemented in the current version.
- Update functions to allow self-fertilization
- Use log on the HMM to avoid underflow computations
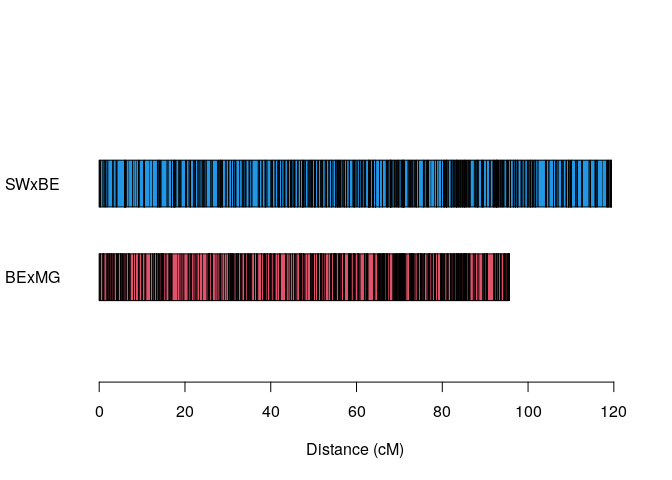
In this section, we will build a consensus map of chromosome 1 using two tetraploid mapping populations: BExMG SWxBE. First, let us load the pre-constructed maps in MAPpoly’s format:
require(mappolymp)
require(mappoly)
map_ch1_BExMG <- readRDS("~/repos/current_work/rose/fullsib_maps/BExMG/map_err_ch_1.rds")
map_ch1_SWxBE <- readRDS("~/repos/current_work/rose/fullsib_maps/SWxBE/map_err_ch_1.rds")
dat_BExMG <- readRDS("~/repos/current_work/rose/data/dat_BExMG.rds")
dat_SWxBE <- readRDS("~/repos/current_work/rose/data/dat_SWxBE.rds")
map.list <- list(BExMG = map_ch1_BExMG,
SWxBE = map_ch1_SWxBE)
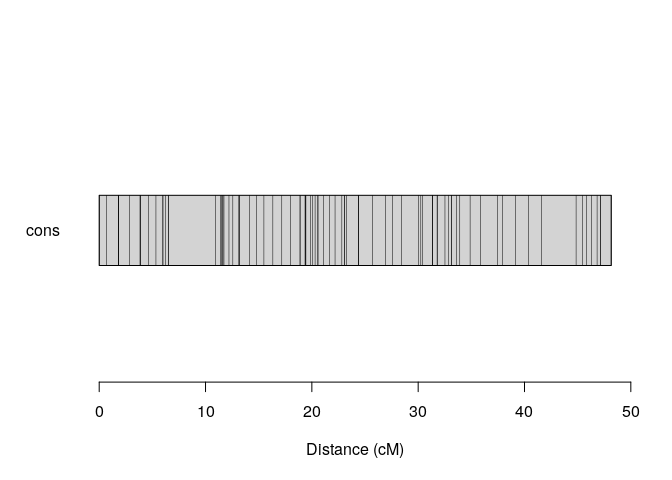
plot_map_list_consensus(map.list, col = c(2,4))Formatting data to MAPpoly2
data.list <- list(BExMG = dat_BExMG,
SWxBE = dat_SWxBE)
parents.mat <- matrix(c("Brite_Eyes","My_Girl",
"Stormy_Weather","Brite_Eyes"),
ncol = 2,
byrow = TRUE,
dimnames = list(c("pop1", "pop2"), c("P1", "P2")))Preparing data and assessing correspondence among homologous chromosomes (holomogs).
w <- prepare_maps_to_merge(map.list = map.list,
data.list = data.list,
parents.mat = parents.mat,
err = 0)## | | | 0% | | | 1% | |= | 1% | |= | 2% | |= | 3% | |== | 3% | |== | 4% | |== | 5% | |=== | 5% | |=== | 6% | |=== | 7% | |==== | 7% | |==== | 8% | |==== | 9% | |===== | 9% | |===== | 10% | |===== | 11% | |====== | 11% | |====== | 12% | |====== | 13% | |======= | 13% | |======= | 14% | |======= | 15% | |======== | 15% | |======== | 16% | |======== | 17% | |========= | 17% | |========= | 18% | |========= | 19% | |========== | 19% | |========== | 20% | |========== | 21% | |=========== | 21% | |=========== | 22% | |=========== | 23% | |============ | 23% | |============ | 24% | |============ | 25% | |============= | 25% | |============= | 26% | |============= | 27% | |============== | 27% | |============== | 28% | |============== | 29% | |=============== | 29% | |=============== | 30% | |=============== | 31% | |================ | 31% | |================ | 32% | |================ | 33% | |================= | 33% | |================= | 34% | |================= | 35% | |================== | 35% | |================== | 36% | |================== | 37% | |=================== | 37% | |=================== | 38% | |=================== | 39% | |==================== | 39% | |==================== | 40% | |==================== | 41% | |===================== | 41% | |===================== | 42% | |===================== | 43% | |====================== | 43% | |====================== | 44% | |====================== | 45% | |======================= | 45% | |======================= | 46% | |======================= | 47% | |======================== | 47% | |======================== | 48% | |======================== | 49% | |========================= | 49% | |========================= | 50% | |========================= | 51% | |========================== | 51% | |========================== | 52% | |========================== | 53% | |=========================== | 53% | |=========================== | 54% | |=========================== | 55% | |============================ | 55% | |============================ | 56% | |============================ | 57% | |============================= | 57% | |============================= | 58% | |============================= | 59% | |============================== | 59% | |============================== | 60% | |============================== | 61% | |=============================== | 61% | |=============================== | 62% | |=============================== | 63% | |================================ | 63% | |================================ | 64% | |================================ | 65% | |================================= | 65% | |================================= | 66% | |================================= | 67% | |================================== | 67% | |================================== | 68% | |================================== | 69% | |=================================== | 69% | |=================================== | 70% | |=================================== | 71% | |==================================== | 71% | |==================================== | 72% | |==================================== | 73% | |===================================== | 73% | |===================================== | 74% | |===================================== | 75% | |====================================== | 75% | |====================================== | 76% | |====================================== | 77% | |======================================= | 77% | |======================================= | 78% | |======================================= | 79% | |======================================== | 79% | |======================================== | 80% | |======================================== | 81% | |========================================= | 81% | |========================================= | 82% | |========================================= | 83% | |========================================== | 83% | |========================================== | 84% | |========================================== | 85% | |=========================================== | 85% | |=========================================== | 86% | |=========================================== | 87% | |============================================ | 87% | |============================================ | 88% | |============================================ | 89% | |============================================= | 89% | |============================================= | 90% | |============================================= | 91% | |============================================== | 91% | |============================================== | 92% | |============================================== | 93% | |=============================================== | 93% | |=============================================== | 94% | |=============================================== | 95% | |================================================ | 95% | |================================================ | 96% | |================================================ | 97% | |================================================= | 97% | |================================================= | 98% | |================================================= | 99% | |==================================================| 99% | |==================================================| 100%
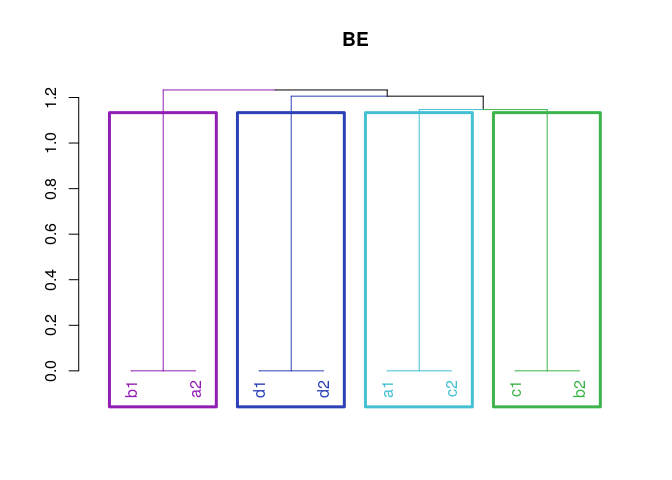
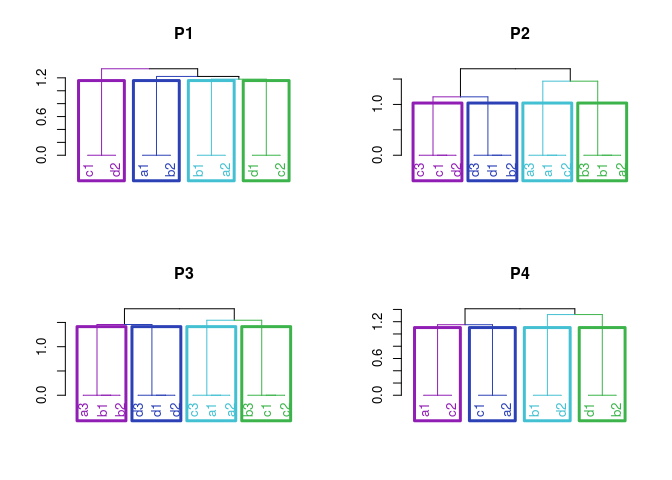
Correspondence among homologous chromosomes in different crosses. In this case, Brite Eyes is present in two crosses. Homolog C in cross 1 (BExMG) corresponds to homolog D in cross 2 (SWxBE). Yet, D corresponds to A, A corresponds to C and B corresponds to B.
plot(w)Map reconstruction (without error - need to fix underflow computations)
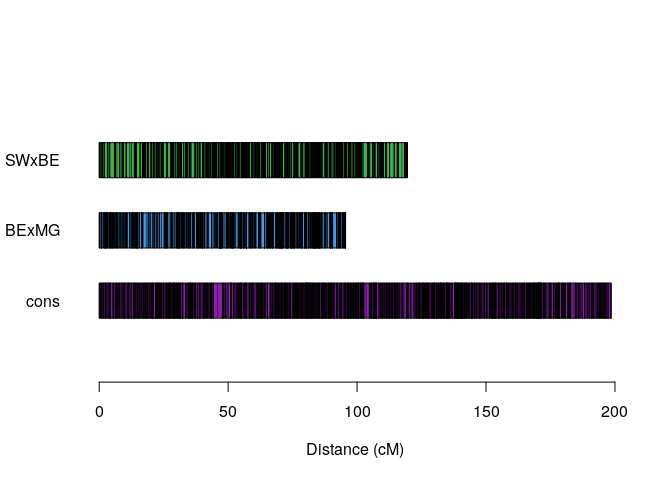
consensus.map <- hmm_map_reconstruction(w, tol = 10e-4, verbose = FALSE)Notice that the map is still longer than expected because we still need to include the genotype errors in the algorithm
plot_map_list_consensus(map.list, consensus.map, col = mp_pallet2(3))Computing homolog probabilities
multi_fam_genoprob <- calc_genoprob_mutli_fam(consensus.map, step = 1)Probability profiles
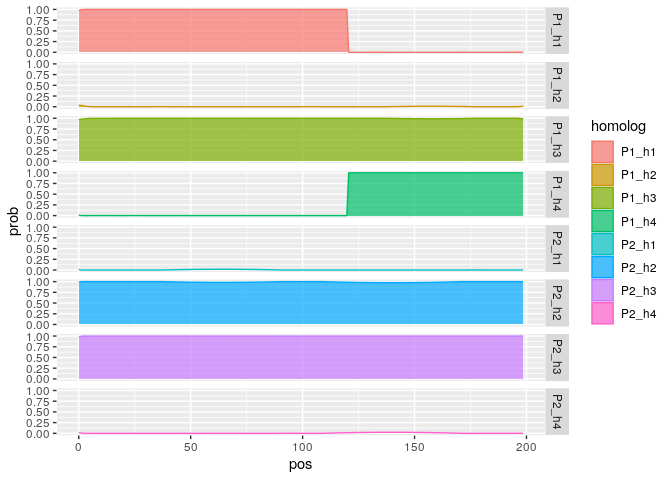
g <- multi_fam_genoprob$genoprob
Z1<- g %>% filter(ind == "X16400_N047")
head(Z1)## ind snp prob pos homolog
## 1 X16400_N047 Affx-86811403 0.98009213 0 P1_h1
## 2 X16400_N047 Affx-86811403 0.03914838 0 P1_h2
## 3 X16400_N047 Affx-86811403 0.96825905 0 P1_h3
## 4 X16400_N047 Affx-86811403 0.01250045 0 P1_h4
## 5 X16400_N047 Affx-86811403 0.01250045 0 P2_h1
## 6 X16400_N047 Affx-86811403 0.98586862 0 P2_h2
ggplot2::ggplot(Z1, ggplot2::aes(x = pos, y = prob, fill = homolog, color = homolog)) +
ggplot2::geom_density(stat = "identity", alpha = 0.7, position = "stack") +
ggplot2::facet_grid(rows = ggplot2::vars(homolog))Let us simulate four parents:
- P1 tetraploid, 50 markers with alleles 0,1,2,3
- P2 diploid, 50 markers with alleles 2,3,4,5
- P3 hexaploid, 50 markers with alleles 0,1,2,3,4,5
- P4 tetraploid, 50 markers with alleles 0,2,6,8
crossed as follows:
- P1 x P2: 100 individuals
- P3 x P1: 100 individuals
- P4 x P2: 100 individuals
- P3 x P4: 100 individuals
map length: 50 cM
ploidy.vec <- c(4, 2, 6, 4) #three parents
names(ploidy.vec) <- c("P1", "P2", "P3", "P4")
cross.mat <- matrix(c("P1","P2",
"P3","P1",
"P4","P2",
"P3","P4"),
ncol = 2, byrow = T)
n.mrk <- c(50,50,50,50) #per parent
map.length <- 50 #in centimorgans
alleles <- list(P1 = c(0:3),
P2 = c(2:5),
P3 = c(0:5),
P4 = c(0,2,6,8))
n.ind <- c(100, 100, 100, 100) #per cross
sim.cross <- simulate_multiple_crosses(ploidy.vec,
cross.mat,
n.ind,
n.mrk,
alleles,
map.length)
sim.cross## This is an object of class mappolymp.data'
## Number of founders: 4
## Ploidy of founders: 4 6 4 2
## No. individuals: 400
## No. markers: 88
##
## Number of individuals per crosses:
## P1 P2 P4
## P1 0 100 0
## P3 100 0 100
## P4 0 100 0
##
## Number of markers per allelic level:
## 2 3 4 5 6 7 8
## 1 5 23 16 16 10 1
w <- states_to_visit(sim.cross)## | | | 0% | | | 1% | |= | 1%
map <- hmm_map_reconstruction(w, tol = 10e-4, verbose = FALSE)plot_map_list_consensus(map.list = NULL, map)Simulating four tetraploid parents with 100 biallelic markers each crossed as follows:
- P1 x P2: 100 individuals
- P1 x P3: 100 individuals
- P2 x P3: 100 individuals
- P2 x P4: 100 individuals
- P3 x P4: 100 individuals
map length: 50 cM
ploidy.vec <- c(4, 4, 4, 4) #four parents
names(ploidy.vec) <- c("P1", "P2", "P3", "P4")
parents.mat <- matrix(c("P1","P2",
"P1","P3",
"P2","P3",
"P2","P4",
"P3","P4"), ncol = 2, byrow = T)
n.mrk <- c(100,100,100,100) #per parent
map.length <- 50 #in centimorgans
alleles <- list(P1 = c(0:1),
P2 = c(0:1),
P3 = c(0:1),
P4 = c(0:1))
n.ind <- c(100, 100, 100, 100, 100)
sim.cross <- simulate_multiple_crosses(ploidy.vec,
parents.mat,
n.ind,
n.mrk,
alleles,
map.length)
sim.cross## This is an object of class mappolymp.data'
## Number of founders: 4
## Ploidy of founders: 4 4 4 4
## No. individuals: 500
## No. markers: 165
##
## Number of individuals per crosses:
## P2 P3 P4
## P1 100 100 0
## P2 0 100 100
## P3 0 0 100
##
## Number of markers per allelic level:
## 1 2
## 7 143
Converting data to MAPpoly
data.list <- mappolymp_to_mappoly(dat = sim.cross)Building full-sib maps
map.list <- vector("list", length(data.list))
names(map.list) <- names(data.list)
for(i in 1:length(data.list)){
dat <- data.list[[i]]
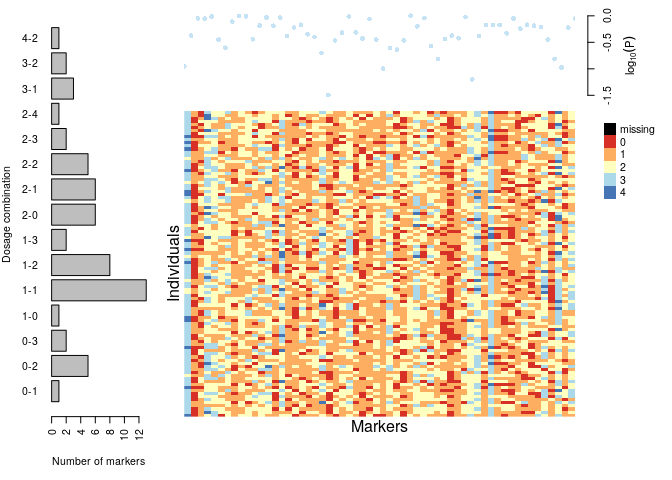
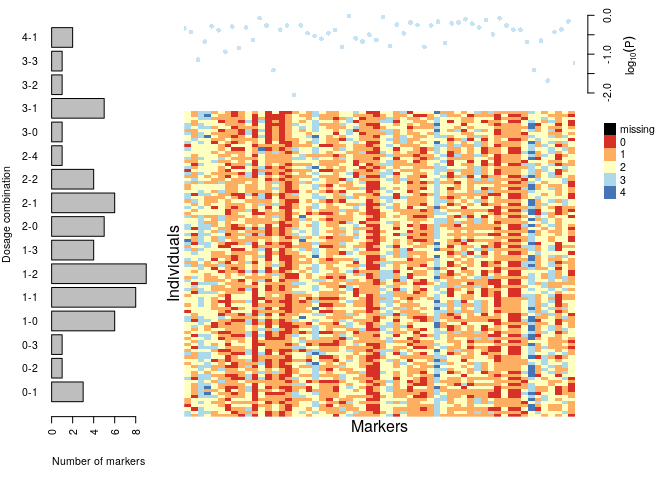
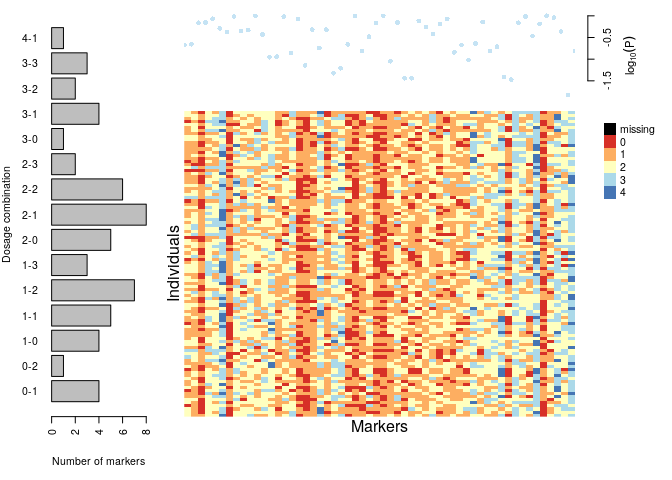
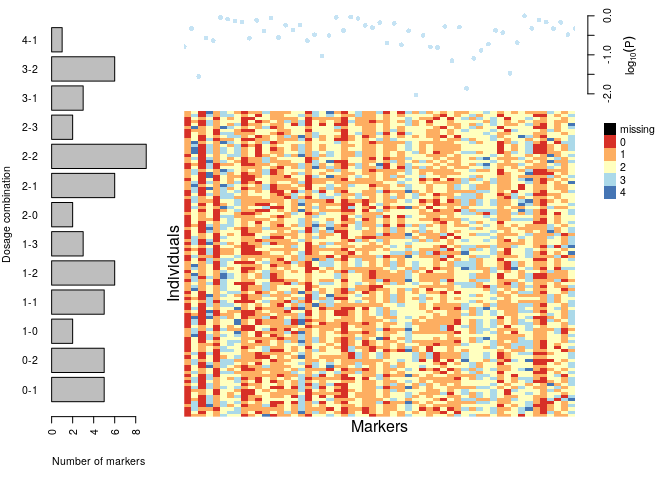
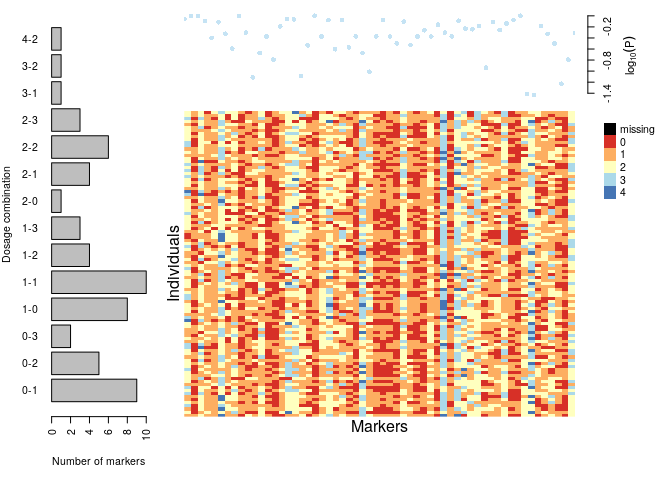
plot(dat)
s <- make_seq_mappoly(dat, "all")
tpt <- est_pairwise_rf(s)
map.list[[i]] <- est_rf_hmm_sequential(s, tpt)
}## INFO: Going singlemode. Using one CPU for calculation.
## Number of markers: 58
## ════════════════════════════════════════════════════════════ Initial sequence ══
## 4 markers...
## • Trying sequence: 1 2 3 4 :
## 20 phase(s): . . . . . . . . . . . . . . . . . . . .
## ══════════════════════════════════════════════════ Done with initial sequence ══
## 5 /5 :(8.6%) 5 : 6 ph (1/6) -- tail: 4 •••| •||| ... •••| ||•|
## 6 /6 :(10.3%) 6 : 1 ph (1/1) -- tail: 5 |•|| ||••
## 7 /7 :(12.1%) 7 : 16 ph (1/16) -- tail: 6 •||| •••| ... •||| ••|•
## 8 /8 :(13.8%) 8 : 1 ph (1/1) -- tail: 7 ||•• ||||
## 9 /9 :(15.5%) 9 : 1 ph (1/1) -- tail: 8 |||| •••|
## 10/10:(17.2%) 10: 1 ph (1/1) -- tail: 9 ||•• •|||
## 11/11:(19%) 11: 2 ph (1/2) -- tail: 10 •|•| |||• •||• |||•
## 12/12:(20.7%) 12: 1 ph (1/1) -- tail: 11 ••|| •|||
## 13/13:(22.4%) 13: 1 ph (1/1) -- tail: 12 ••|| ||••
## 14/14:(24.1%) 14: 1 ph (1/1) -- tail: 13 ||•| ||•|
## 15/15:(25.9%) 15: 1 ph (1/1) -- tail: 14 •||• |•••
## 16/16:(27.6%) 16: 4 ph (1/4) -- tail: 15 •|•| |||| ... •||• ||||
## 17/17:(29.3%) 17: 1 ph (1/1) -- tail: 16 |•|| |||•
## 18/18:(31%) 18: 1 ph (1/1) -- tail: 17 |||• |••|
## 19/19:(32.8%) 19: 1 ph (1/1) -- tail: 18 |••| ||||
## 20/20:(34.5%) 20: 1 ph (1/1) -- tail: 19 ||•| ||••
## 21/21:(36.2%) 21: 1 ph (1/1) -- tail: 20 ••|| |••|
## 22/22:(37.9%) 22: 1 ph (1/1) -- tail: 21 |||| |••|
## 23/23:(39.7%) 23: 1 ph (1/1) -- tail: 22 |•|| |||•
## 24/24:(41.4%) 24: 1 ph (1/1) -- tail: 23 •||| ||••
## 25/25:(43.1%) 25: 1 ph (1/1) -- tail: 24 ••|• |•||
## 26/26:(44.8%) 26: 1 ph (1/1) -- tail: 25 ||•| |||•
## 27/27:(46.6%) 27: 2 ph (1/2) -- tail: 26 |||• |•|| |||• ||•|
## 28/28:(48.3%) 28: 1 ph (1/1) -- tail: 27 •|•| ||||
## 29/29:(50%) 29: 1 ph (1/1) -- tail: 28 |•|| ||••
## 30/30:(51.7%) 30: 1 ph (1/1) -- tail: 29 |||| ||••
## 31/31:(53.4%) 31: 1 ph (1/1) -- tail: 30 ••|• ||•|
## 32/32:(55.2%) 32: 1 ph (1/1) -- tail: 31 •||| •|||
## 33/33:(56.9%) 33: 1 ph (1/1) -- tail: 32 |•|| ||•|
## 34/34:(58.6%) 34: 1 ph (1/1) -- tail: 33 |||| •••|
## 35/35:(60.3%) 35: 1 ph (1/1) -- tail: 34 •||• ||••
## 36/36:(62.1%) 36: 1 ph (1/1) -- tail: 35 |••| •|||
## 37/37:(63.8%) 37: 1 ph (1/1) -- tail: 36 •||• |||•
## 38/38:(65.5%) 38: 1 ph (1/1) -- tail: 37 |||• •|•|
## 39/39:(67.2%) 39: 1 ph (1/1) -- tail: 38 |||| •||•
## 40/40:(69%) 40: 1 ph (1/1) -- tail: 39 |•|| ||||
## 41/41:(70.7%) 41: 1 ph (1/1) -- tail: 40 |||| •|•|
## 42/42:(72.4%) 42: 1 ph (1/1) -- tail: 41 ||•• |||•
## 43/43:(74.1%) 43: 1 ph (1/1) -- tail: 42 ••|| ••||
## 44/44:(75.9%) 44: 1 ph (1/1) -- tail: 43 |••• ||••
## 45/45:(77.6%) 45: 1 ph (1/1) -- tail: 44 |||| •|•|
## 46/46:(79.3%) 46: 1 ph (1/1) -- tail: 45 |••| ••••
## 47/47:(81%) 47: 1 ph (1/1) -- tail: 46 •||• ||||
## 48/48:(82.8%) 48: 1 ph (1/1) -- tail: 47 |||• |||•
## 49/49:(84.5%) 49: 1 ph (1/1) -- tail: 48 |||• •|||
## 50/50:(86.2%) 50: 1 ph (1/1) -- tail: 49 ••|| ||||
## 51/51:(87.9%) 51: 1 ph (1/1) -- tail: 50 |•|| |••|
## 52/52:(89.7%) 52: 2 ph (1/2) -- tail: 51 |||• |•|| |||• ||•|
## 53/53:(91.4%) 53: 1 ph (1/1) -- tail: 52 ||•| ••||
## 54/54:(93.1%) 54: 1 ph (1/1) -- tail: 53 •|•| ||••
## 55/55:(94.8%) 55: 2 ph (1/2) -- tail: 54 |•|| •||| |•|| |•||
## 56/56:(96.6%) 56: 1 ph (1/1) -- tail: 55 •|•• |••|
## 57/57:(98.3%) 57: 1 ph (1/1) -- tail: 56 •||| ||•|
## 58/58:(100%) 58: 1 ph (1/1) -- tail: 57 •||| |•••
## ══════════════════════════════════ Reestimating final recombination fractions ══
## Markers in the initial sequence: 58
## Mapped markers : 58 (100%)
## ════════════════════════════════════════════════════════════════════════════════
## INFO: Going singlemode. Using one CPU for calculation.
## Number of markers: 58
## ════════════════════════════════════════════════════════════ Initial sequence ══
## 4 markers...
## • Trying sequence: 1 2 3 4 :
## 64 phase(s): . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
## . . . . . . . . . . . . . . . . . . . . . . . . . .
## ══════════════════════════════════════════════════ Done with initial sequence ══
## 5 /5 :(8.6%) 5 : 12 ph (2/12) -- tail: 4 ••|| |•|| ... ••|| |||•
## 6 /6 :(10.3%) 6 : 8 ph (1/8) -- tail: 5 •||| ••|| ... •||| |••|
## 7 /7 :(12.1%) 7 : 4 ph (1/4) -- tail: 6 •||| •||| ... •||| |•||
## 8 /8 :(13.8%) 8 : 1 ph (1/1) -- tail: 7 |||• ||•|
## 9 /9 :(15.5%) 9 : 1 ph (1/1) -- tail: 8 ||•• ||||
## 10/10:(17.2%) 10: 1 ph (1/1) -- tail: 9 |•|| |•••
## 11/11:(19%) 11: 1 ph (1/1) -- tail: 10 |||| •|||
## 12/12:(20.7%) 12: 2 ph (1/2) -- tail: 11 ••|| ••|| ••|| •||•
## 13/13:(22.4%) 13: 1 ph (1/1) -- tail: 12 |||• ||||
## 14/14:(24.1%) 14: 1 ph (1/1) -- tail: 13 |•|• •|||
## 15/15:(25.9%) 15: 1 ph (1/1) -- tail: 14 |||• ||||
## 16/16:(27.6%) 16: 1 ph (1/1) -- tail: 15 •||| ||||
## 17/17:(29.3%) 17: 1 ph (1/1) -- tail: 16 •||| |•|•
## 18/18:(31%) 18: 1 ph (1/1) -- tail: 17 •||• ••||
## 19/19:(32.8%) 19: 1 ph (1/1) -- tail: 18 •||| •||•
## 20/20:(34.5%) 20: 1 ph (1/1) -- tail: 19 •••| •||•
## 21/21:(36.2%) 21: 1 ph (1/1) -- tail: 20 ••|| •|||
## 22/22:(37.9%) 22: 1 ph (1/1) -- tail: 21 ••|| ||||
## 23/23:(39.7%) 23: 1 ph (1/1) -- tail: 22 |||| •||•
## 24/24:(41.4%) 24: 1 ph (1/1) -- tail: 23 |•|| |•|•
## 25/25:(43.1%) 25: 1 ph (1/1) -- tail: 24 •••| ||•|
## 26/26:(44.8%) 26: 1 ph (1/1) -- tail: 25 ||•| ••||
## 27/27:(46.6%) 27: 1 ph (1/1) -- tail: 26 |•|• •|||
## 28/28:(48.3%) 28: 1 ph (1/1) -- tail: 27 •||| ||||
## 29/29:(50%) 29: 1 ph (1/1) -- tail: 28 |||| ||•|
## 30/30:(51.7%) 30: 1 ph (1/1) -- tail: 29 |•|| •|••
## 31/31:(53.4%) 31: 1 ph (1/1) -- tail: 30 •••• ||•|
## 32/32:(55.2%) 32: 1 ph (1/1) -- tail: 31 ||•• ||||
## 33/33:(56.9%) 33: 1 ph (1/1) -- tail: 32 •••| •|||
## 34/34:(58.6%) 34: 1 ph (1/1) -- tail: 33 •||| |||•
## 35/35:(60.3%) 35: 1 ph (1/1) -- tail: 34 |••| ||||
## 36/36:(62.1%) 36: 1 ph (1/1) -- tail: 35 ||•| ||•|
## 37/37:(63.8%) 37: 1 ph (1/1) -- tail: 36 |||| •|••
## 38/38:(65.5%) 38: 1 ph (1/1) -- tail: 37 |••| ••••
## 39/39:(67.2%) 39: 2 ph (1/2) -- tail: 38 ••|• |||• •|•• |||•
## 40/40:(69%) 40: 1 ph (1/1) -- tail: 39 ||•| |•||
## 41/41:(70.7%) 41: 1 ph (1/1) -- tail: 40 ||•| ||••
## 42/42:(72.4%) 42: 2 ph (1/2) -- tail: 41 •||• •||| •||• |•||
## 43/43:(74.1%) 43: 1 ph (1/1) -- tail: 42 |••| ||||
## 44/44:(75.9%) 44: 1 ph (1/1) -- tail: 43 ||•| •|••
## 45/45:(77.6%) 45: 1 ph (1/1) -- tail: 44 ||•| |•||
## 46/46:(79.3%) 46: 1 ph (1/1) -- tail: 45 ||•| •|•|
## 47/47:(81%) 47: 1 ph (1/1) -- tail: 46 |||| •|||
## 48/48:(82.8%) 48: 1 ph (1/1) -- tail: 47 ••|• ||||
## 49/49:(84.5%) 49: 1 ph (1/1) -- tail: 48 ||•| ||||
## 50/50:(86.2%) 50: 1 ph (1/1) -- tail: 49 ||•| ||||
## 51/51:(87.9%) 51: 2 ph (1/2) -- tail: 50 ••|| |||• |•|• |||•
## 52/52:(89.7%) 52: 1 ph (1/1) -- tail: 51 |••• ••|•
## 53/53:(91.4%) 53: 1 ph (1/1) -- tail: 52 •||| •|•|
## 54/54:(93.1%) 54: 1 ph (1/1) -- tail: 53 •|•• |||•
## 55/55:(94.8%) 55: 1 ph (1/1) -- tail: 54 |•|| •|••
## 56/56:(96.6%) 56: 1 ph (1/1) -- tail: 55 •||| •|||
## 57/57:(98.3%) 57: 1 ph (1/1) -- tail: 56 •••| •|||
## 58/58:(100%) 58: 1 ph (1/1) -- tail: 57 ||•| |||•
## ══════════════════════════════════ Reestimating final recombination fractions ══
## Markers in the initial sequence: 58
## Mapped markers : 58 (100%)
## ════════════════════════════════════════════════════════════════════════════════
## INFO: Going singlemode. Using one CPU for calculation.
## Number of markers: 56
## ════════════════════════════════════════════════════════════ Initial sequence ══
## 4 markers...
## • Trying sequence: 1 2 3 4 :
## 4 phase(s): . . . .
## ══════════════════════════════════════════════════ Done with initial sequence ══
## 5 /5 :(8.9%) 5 : 1 ph (1/1) -- tail: 4 •••| •|||
## 6 /6 :(10.7%) 6 : 3 ph (1/3) -- tail: 5 •••| •••| ... ••|• •••|
## 7 /7 :(12.5%) 7 : 1 ph (1/1) -- tail: 6 |•|| ||||
## 8 /8 :(14.3%) 8 : 3 ph (1/3) -- tail: 7 |•|• |••| ... |•|• |•|•
## 9 /9 :(16.1%) 9 : 12 ph (1/12) -- tail: 8 •|•| •||| ... •|•| |•||
## 10/10:(17.9%) 10: 1 ph (1/1) -- tail: 9 |••• |•||
## 11/11:(19.6%) 11: 1 ph (1/1) -- tail: 10 •||• |||•
## 12/12:(21.4%) 12: 4 ph (1/4) -- tail: 11 •••| •||| ... •••| ||•|
## 13/13:(23.2%) 13: 1 ph (1/1) -- tail: 12 ||•• ||••
## 14/14:(25%) 14: 1 ph (1/1) -- tail: 13 |•|• ||||
## 15: not included (*linkage phases*)
## 15/16:(28.6%) 16: 6 ph (1/6) -- tail: 14 •||• ••|| ... •||• •||•
## 16/17:(30.4%) 17: 1 ph (1/1) -- tail: 15 |||| ||•|
## 17/18:(32.1%) 18: 1 ph (1/1) -- tail: 16 |||• ||||
## 18/19:(33.9%) 19: 1 ph (1/1) -- tail: 17 |||| •|•|
## 19/20:(35.7%) 20: 2 ph (1/2) -- tail: 18 •|•• |••| •|•• ||••
## 20/21:(37.5%) 21: 1 ph (1/1) -- tail: 19 |••| ||||
## 21/22:(39.3%) 22: 1 ph (1/1) -- tail: 20 |••| ||••
## 22/23:(41.1%) 23: 3 ph (1/3) -- tail: 21 •||• •||• ... |•|• •||•
## 23/24:(42.9%) 24: 1 ph (1/1) -- tail: 22 |•|| |•||
## 24/25:(44.6%) 25: 1 ph (1/1) -- tail: 23 |||| ||•|
## 25/26:(46.4%) 26: 1 ph (1/1) -- tail: 24 |•|• ||||
## 26/27:(48.2%) 27: 2 ph (1/2) -- tail: 25 ||•| •||• |||• •||•
## 27/28:(50%) 28: 1 ph (1/1) -- tail: 26 ||•| ||||
## 28/29:(51.8%) 29: 1 ph (1/1) -- tail: 27 |||| ||•|
## 29/30:(53.6%) 30: 1 ph (1/1) -- tail: 28 ||•• ||||
## 30/31:(55.4%) 31: 1 ph (1/1) -- tail: 29 |••| |||•
## 31/32:(57.1%) 32: 1 ph (1/1) -- tail: 30 |||• •|||
## 32/33:(58.9%) 33: 2 ph (1/2) -- tail: 31 •|•| |•|| •|•| ||•|
## 33/34:(60.7%) 34: 1 ph (1/1) -- tail: 32 ||•| ||•|
## 34/35:(62.5%) 35: 1 ph (1/1) -- tail: 33 ••|• ||||
## 35/36:(64.3%) 36: 1 ph (1/1) -- tail: 34 ||•| •|•|
## 36/37:(66.1%) 37: 1 ph (1/1) -- tail: 35 ||•| |||•
## 37/38:(67.9%) 38: 1 ph (1/1) -- tail: 36 |||• |•|•
## 38/39:(69.6%) 39: 1 ph (1/1) -- tail: 37 ||•• ||||
## 39/40:(71.4%) 40: 1 ph (1/1) -- tail: 38 •||| ••||
## 40/41:(73.2%) 41: 1 ph (1/1) -- tail: 39 •|•| |•||
## 41/42:(75%) 42: 1 ph (1/1) -- tail: 40 •||• |•••
## 42/43:(76.8%) 43: 1 ph (1/1) -- tail: 41 ||•• |||•
## 43/44:(78.6%) 44: 1 ph (1/1) -- tail: 42 ••|• |•||
## 44/45:(80.4%) 45: 1 ph (1/1) -- tail: 43 |•|| |•••
## 45/46:(82.1%) 46: 1 ph (1/1) -- tail: 44 |••• •||•
## 46/47:(83.9%) 47: 1 ph (1/1) -- tail: 45 •||| |•||
## 47/48:(85.7%) 48: 1 ph (1/1) -- tail: 46 •••• ||•|
## 48/49:(87.5%) 49: 1 ph (1/1) -- tail: 47 |||• |•••
## 49/50:(89.3%) 50: 1 ph (1/1) -- tail: 48 •||| |••|
## 50/51:(91.1%) 51: 1 ph (1/1) -- tail: 49 ••|• •|••
## 51/52:(92.9%) 52: 1 ph (1/1) -- tail: 50 |•|| ||||
## 52/53:(94.6%) 53: 1 ph (1/1) -- tail: 51 ||•• |||•
## 53/54:(96.4%) 54: 1 ph (1/1) -- tail: 52 |••| •|••
## 54/55:(98.2%) 55: 1 ph (1/1) -- tail: 53 •|•| ••||
## 55/56:(100%) 56: 2 ph (1/2) -- tail: 54 ••|• |••• |••• |•••
## ══════════════════════════════════ Reestimating final recombination fractions ══
## Markers in the initial sequence: 56
## Mapped markers : 55 (98.2%)
## ════════════════════════════════════════════════════════════════════════════════
## INFO: Going singlemode. Using one CPU for calculation.
## Number of markers: 55
## ════════════════════════════════════════════════════════════ Initial sequence ══
## 4 markers...
## • Trying sequence: 1 2 3 4 :
## 8 phase(s): . . . . . . . .
## ══════════════════════════════════════════════════ Done with initial sequence ══
## 5 /5 :(9.1%) 5 : 3 ph (1/3) -- tail: 4 •||| |||| ... |•|| ||||
## 6 /6 :(10.9%) 6 : 4 ph (1/4) -- tail: 5 ••|| •••| ... ••|| ••|•
## 7 /7 :(12.7%) 7 : 6 ph (1/6) -- tail: 6 •••| •||| ... •••| |•||
## 8 /8 :(14.5%) 8 : 20 ph (1/20) -- tail: 7 •|•| ••|| ... •|•| •||•
## 9 /9 :(16.4%) 9 : 1 ph (1/1) -- tail: 8 |||| |•||
## 10/10:(18.2%) 10: 1 ph (1/1) -- tail: 9 |•|| |•||
## 11/11:(20%) 11: 2 ph (1/2) -- tail: 10 •||| •||| •||| ||•|
## 12/12:(21.8%) 12: 2 ph (1/2) -- tail: 11 ••|| •|•| ••|| |••|
## 13/13:(23.6%) 13: 1 ph (1/1) -- tail: 12 |||• |||•
## 14/14:(25.5%) 14: 2 ph (1/2) -- tail: 13 |||| •|•| |||| •||•
## 15/15:(27.3%) 15: 1 ph (1/1) -- tail: 14 |•|| |•|•
## 16/16:(29.1%) 16: 1 ph (1/1) -- tail: 15 •||• •|||
## 17/17:(30.9%) 17: 1 ph (1/1) -- tail: 16 |••• |•|•
## 18/18:(32.7%) 18: 1 ph (1/1) -- tail: 17 |||| |•||
## 19/19:(34.5%) 19: 2 ph (1/2) -- tail: 18 ••|| |••| |•|• |••|
## 20/20:(36.4%) 20: 1 ph (1/1) -- tail: 19 •||• •|||
## 21/21:(38.2%) 21: 1 ph (1/1) -- tail: 20 |•|• •|•|
## 22/22:(40%) 22: 1 ph (1/1) -- tail: 21 •||| ••||
## 23/23:(41.8%) 23: 1 ph (1/1) -- tail: 22 |||| •|||
## 24/24:(43.6%) 24: 1 ph (1/1) -- tail: 23 ••|| ||•|
## 25/25:(45.5%) 25: 1 ph (1/1) -- tail: 24 |||• •••|
## 26/26:(47.3%) 26: 1 ph (1/1) -- tail: 25 |||| •|•|
## 27/27:(49.1%) 27: 1 ph (1/1) -- tail: 26 ||•• ||||
## 28/28:(50.9%) 28: 1 ph (1/1) -- tail: 27 ||•• |••|
## 29/29:(52.7%) 29: 1 ph (1/1) -- tail: 28 |||| •||•
## 30/30:(54.5%) 30: 1 ph (1/1) -- tail: 29 |||• •|••
## 31/31:(56.4%) 31: 1 ph (1/1) -- tail: 30 ||•| •||•
## 32/32:(58.2%) 32: 1 ph (1/1) -- tail: 31 •••| |••|
## 33/33:(60%) 33: 1 ph (1/1) -- tail: 32 •|•• •|||
## 34/34:(61.8%) 34: 1 ph (1/1) -- tail: 33 |•|• ||•|
## 35/35:(63.6%) 35: 1 ph (1/1) -- tail: 34 ||•• •|||
## 36/36:(65.5%) 36: 1 ph (1/1) -- tail: 35 |•|| ||•|
## 37/37:(67.3%) 37: 1 ph (1/1) -- tail: 36 •|•| ||•|
## 38/38:(69.1%) 38: 1 ph (1/1) -- tail: 37 |•|• ||||
## 39/39:(70.9%) 39: 1 ph (1/1) -- tail: 38 •||| •|•|
## 40/40:(72.7%) 40: 1 ph (1/1) -- tail: 39 •|•• •||•
## 41/41:(74.5%) 41: 1 ph (1/1) -- tail: 40 ••|• •|||
## 42/42:(76.4%) 42: 1 ph (1/1) -- tail: 41 ||•| |•••
## 43/43:(78.2%) 43: 1 ph (1/1) -- tail: 42 ||•• |••|
## 44/44:(80%) 44: 1 ph (1/1) -- tail: 43 •••• |||•
## 45/45:(81.8%) 45: 1 ph (1/1) -- tail: 44 |||| ||••
## 46/46:(83.6%) 46: 1 ph (1/1) -- tail: 45 |•|| |•||
## 47/47:(85.5%) 47: 1 ph (1/1) -- tail: 46 •||• •||•
## 48/48:(87.3%) 48: 1 ph (1/1) -- tail: 47 ||•• |•|•
## 49/49:(89.1%) 49: 1 ph (1/1) -- tail: 48 •|•| ••|•
## 50/50:(90.9%) 50: 1 ph (1/1) -- tail: 49 |||| ||••
## 51/51:(92.7%) 51: 1 ph (1/1) -- tail: 50 |||| ||•|
## 52/52:(94.5%) 52: 1 ph (1/1) -- tail: 51 |•|• ||••
## 53/53:(96.4%) 53: 1 ph (1/1) -- tail: 52 ||•| |••|
## 54/54:(98.2%) 54: 1 ph (1/1) -- tail: 53 |||• •||•
## 55/55:(100%) 55: 1 ph (1/1) -- tail: 54 |••• •||•
## ══════════════════════════════════ Reestimating final recombination fractions ══
## Markers in the initial sequence: 55
## Mapped markers : 55 (100%)
## ════════════════════════════════════════════════════════════════════════════════
## INFO: Going singlemode. Using one CPU for calculation.
## Number of markers: 58
## ════════════════════════════════════════════════════════════ Initial sequence ══
## 4 markers...
## • Trying sequence: 1 2 3 4 :
## 12 phase(s): . . . . . . . . . . . .
## ══════════════════════════════════════════════════ Done with initial sequence ══
## 5 /5 :(8.6%) 5 : 3 ph (3/3) -- tail: 4 |||| ||•• ... |||| |••|
## 6 /6 :(10.3%) 6 : 5 ph (3/5) -- tail: 5 ||•• •|•• ... ||•• •|••
## 7 /7 :(12.1%) 7 : 4 ph (1/4) -- tail: 6 ||•| ||•| ... ||•| |||•
## 8 /8 :(13.8%) 8 : 12 ph (1/12) -- tail: 7 •||| ••|| ... •||| •||•
## 9 /9 :(15.5%) 9 : 1 ph (1/1) -- tail: 8 |||| |||•
## 10/10:(17.2%) 10: 1 ph (1/1) -- tail: 9 |||| •|•|
## 11/11:(19%) 11: 2 ph (1/2) -- tail: 10 •||| •||| ||•| •|||
## 12/12:(20.7%) 12: 1 ph (1/1) -- tail: 11 ||•• |•||
## 13/13:(22.4%) 13: 1 ph (1/1) -- tail: 12 |||| •|||
## 14/14:(24.1%) 14: 1 ph (1/1) -- tail: 13 •||| ||||
## 15/15:(25.9%) 15: 1 ph (1/1) -- tail: 14 |||| |•|•
## 16/16:(27.6%) 16: 1 ph (1/1) -- tail: 15 |•|• |•|•
## 17/17:(29.3%) 17: 1 ph (1/1) -- tail: 16 •|•| |•|•
## 18/18:(31%) 18: 5 ph (1/5) -- tail: 17 ••|| ||•| ... •|•| ||•|
## 19/19:(32.8%) 19: 1 ph (1/1) -- tail: 18 •||| |||•
## 20/20:(34.5%) 20: 1 ph (1/1) -- tail: 19 |||| ||•|
## 21/21:(36.2%) 21: 1 ph (1/1) -- tail: 20 |||| •••|
## 22/22:(37.9%) 22: 1 ph (1/1) -- tail: 21 |•|• •|•|
## 23/23:(39.7%) 23: 1 ph (1/1) -- tail: 22 ||•| |•||
## 24/24:(41.4%) 24: 1 ph (1/1) -- tail: 23 ||•| ||••
## 25/25:(43.1%) 25: 1 ph (1/1) -- tail: 24 •||| ||•|
## 26/26:(44.8%) 26: 1 ph (1/1) -- tail: 25 |||| •|||
## 27/27:(46.6%) 27: 1 ph (1/1) -- tail: 26 |•|• •|••
## 28/28:(48.3%) 28: 1 ph (1/1) -- tail: 27 •||| •|•|
## 29/29:(50%) 29: 1 ph (1/1) -- tail: 28 ||•| ||||
## 30/30:(51.7%) 30: 1 ph (1/1) -- tail: 29 |||| ||•|
## 31/31:(53.4%) 31: 1 ph (1/1) -- tail: 30 ||•| ||||
## 32/32:(55.2%) 32: 1 ph (1/1) -- tail: 31 |||| •|||
## 33/33:(56.9%) 33: 1 ph (1/1) -- tail: 32 •||| •••|
## 34/34:(58.6%) 34: 1 ph (1/1) -- tail: 33 |||| •||•
## 35/35:(60.3%) 35: 1 ph (1/1) -- tail: 34 |||| •|||
## 36/36:(62.1%) 36: 1 ph (1/1) -- tail: 35 ||•| ||||
## 37/37:(63.8%) 37: 1 ph (1/1) -- tail: 36 |||| |•••
## 38/38:(65.5%) 38: 2 ph (1/2) -- tail: 37 |•|| |||| |||• ||||
## 39/39:(67.2%) 39: 1 ph (1/1) -- tail: 38 •••• •|•|
## 40/40:(69%) 40: 2 ph (1/2) -- tail: 39 |•|| |||| |||• ||||
## 41/41:(70.7%) 41: 1 ph (1/1) -- tail: 40 •|•• •|•|
## 42/42:(72.4%) 42: 1 ph (1/1) -- tail: 41 ••|| |•|•
## 43/43:(74.1%) 43: 1 ph (1/1) -- tail: 42 |•|• ||•|
## 44/44:(75.9%) 44: 1 ph (1/1) -- tail: 43 ||•| ••|•
## 45/45:(77.6%) 45: 1 ph (1/1) -- tail: 44 •||| |•||
## 46/46:(79.3%) 46: 1 ph (1/1) -- tail: 45 |||| ••||
## 47/47:(81%) 47: 1 ph (1/1) -- tail: 46 |•|| •|||
## 48/48:(82.8%) 48: 1 ph (1/1) -- tail: 47 •|•• |||•
## 49/49:(84.5%) 49: 1 ph (1/1) -- tail: 48 |•|| ||||
## 50/50:(86.2%) 50: 1 ph (1/1) -- tail: 49 |||| |•||
## 51/51:(87.9%) 51: 1 ph (1/1) -- tail: 50 |||• ••||
## 52/52:(89.7%) 52: 1 ph (1/1) -- tail: 51 |•|• |•••
## 53/53:(91.4%) 53: 1 ph (1/1) -- tail: 52 •||| |||•
## 54/54:(93.1%) 54: 2 ph (1/2) -- tail: 53 ||•| |||• |||• |||•
## 55/55:(94.8%) 55: 1 ph (1/1) -- tail: 54 |•|• ||•|
## 56/56:(96.6%) 56: 1 ph (1/1) -- tail: 55 •||| ||•|
## 57/57:(98.3%) 57: 1 ph (1/1) -- tail: 56 •||| ||||
## 58/58:(100%) 58: 1 ph (1/1) -- tail: 57 |||• ••|•
## ══════════════════════════════════ Reestimating final recombination fractions ══
## Markers in the initial sequence: 58
## Mapped markers : 58 (100%)
## ════════════════════════════════════════════════════════════════════════════════
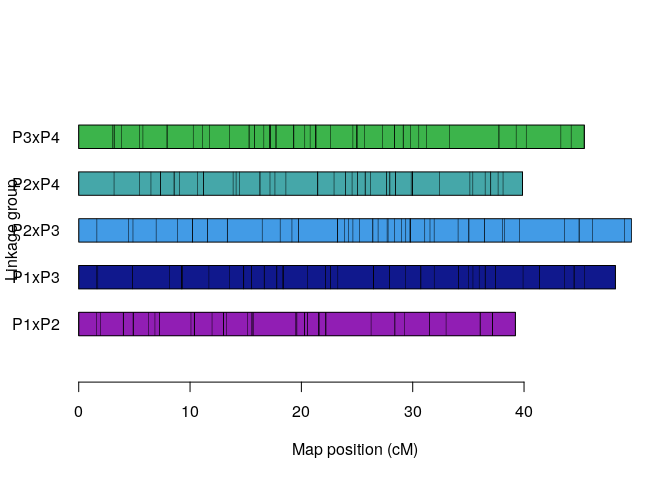
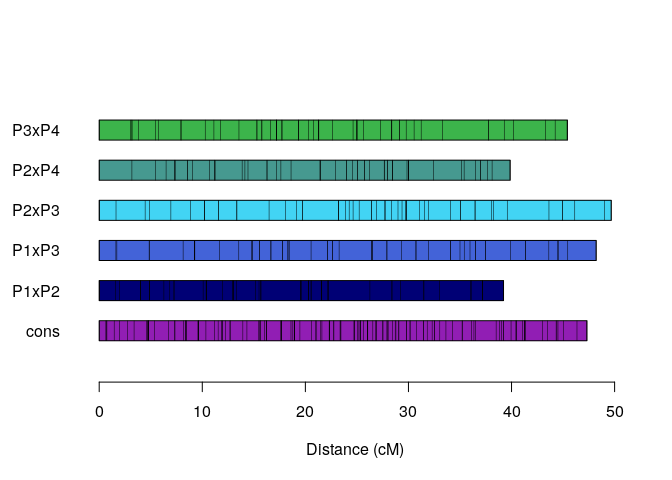
plot_map_list(map.list, col = mp_pallet2(5))Preparing data and assessing correspondence among homologous chromosomes (holomogs).
w <- prepare_maps_to_merge(map.list = map.list,
data.list = data.list,
parents.mat = parents.mat,
err = 0)## | | | 0% | | | 1% | |= | 1% | |= | 2% | |= | 3% | |== | 3% | |== | 4% | |== | 5% | |=== | 6% | |=== | 7% | |==== | 8% | |===== | 9% | |===== | 10% | |====== | 11% | |====== | 12% | |====== | 13% | |======= | 13% | |======= | 14% | |======= | 15% | |======== | 15% | |======== | 16% | |======== | 17% | |========= | 17% | |========= | 18% | |========= | 19% | |========== | 20% | |========== | 21% | |=========== | 22% | |============ | 23% | |============ | 24% | |============= | 25% | |============= | 26% | |============= | 27% | |============== | 27% | |============== | 28% | |============== | 29% | |=============== | 29% | |=============== | 30% | |=============== | 31% | |================ | 31% | |================ | 32% | |================ | 33% | |================= | 34% | |================= | 35% | |================== | 36% | |=================== | 37% | |=================== | 38% | |==================== | 39% | |==================== | 40% | |==================== | 41% | |===================== | 41% | |===================== | 42% | |===================== | 43% | |====================== | 43% | |====================== | 44% | |====================== | 45% | |======================= | 45% | |======================= | 46% | |======================= | 47% | |======================== | 48% | |======================== | 49% | |========================= | 50% | |========================== | 51% | |========================== | 52% | |=========================== | 53% | |=========================== | 54% | |=========================== | 55% | |============================ | 55% | |============================ | 56% | |============================ | 57% | |============================= | 57% | |============================= | 58% | |============================= | 59% | |============================== | 59% | |============================== | 60% | |============================== | 61% | |=============================== | 62% | |=============================== | 63% | |================================ | 64% | |================================= | 65% | |================================= | 66% | |================================== | 67% | |================================== | 68% | |================================== | 69% | |=================================== | 69% | |=================================== | 70% | |=================================== | 71% | |==================================== | 71% | |==================================== | 72% | |==================================== | 73% | |===================================== | 73% | |===================================== | 74% | |===================================== | 75% | |====================================== | 76% | |====================================== | 77% | |======================================= | 78% | |======================================== | 79% | |======================================== | 80% | |========================================= | 81% | |========================================= | 82% | |========================================= | 83% | |========================================== | 83% | |========================================== | 84% | |========================================== | 85% | |=========================================== | 85% | |=========================================== | 86% | |=========================================== | 87% | |============================================ | 87% | |============================================ | 88% | |============================================ | 89% | |============================================= | 90% | |============================================= | 91% | |============================================== | 92% | |=============================================== | 93% | |=============================================== | 94% | |================================================ | 95% | |================================================ | 96% | |================================================ | 97% | |================================================= | 97% | |================================================= | 98% | |================================================= | 99% | |==================================================| 99% | |==================================================| 100%
Correspondence among homologous chromosomes in different crosses.
plot(w)consensus.map <- hmm_map_reconstruction(w, tol = 10e-4, verbose = FALSE)plot_map_list_consensus(map.list, consensus.map, col = mp_pallet2(6))multi_fam_genoprob <- calc_genoprob_mutli_fam(consensus.map, step = 1)g <- multi_fam_genoprob$genoprob
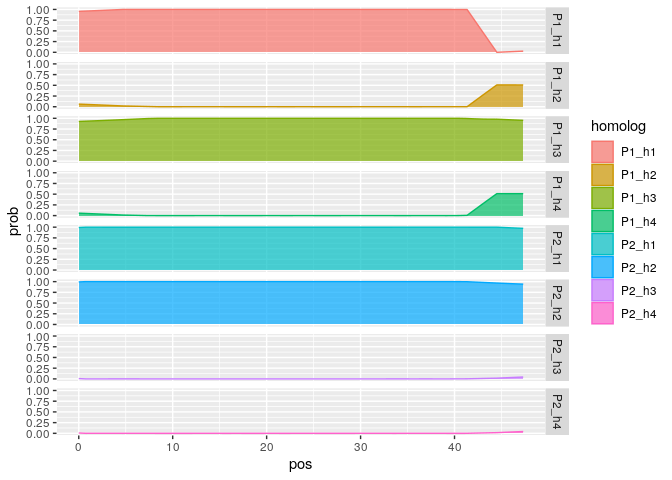
Z1<- g %>% filter(ind == unique(g$ind)[1])
head(Z1)## ind snp prob pos homolog
## 1 Ind_P1xP2_1 M1 0.95512461 0 P1_h1
## 2 Ind_P1xP2_1 M1 0.06143484 0 P1_h2
## 3 Ind_P1xP2_1 M1 0.92662983 0 P1_h3
## 4 Ind_P1xP2_1 M1 0.05681072 0 P1_h4
## 5 Ind_P1xP2_1 M1 0.99258438 0 P2_h1
## 6 Ind_P1xP2_1 M1 0.99258438 0 P2_h2
ggplot2::ggplot(Z1, ggplot2::aes(x = pos, y = prob, fill = homolog, color = homolog)) +
ggplot2::geom_density(stat = "identity", alpha = 0.7, position = "stack") +
ggplot2::facet_grid(rows = ggplot2::vars(homolog))