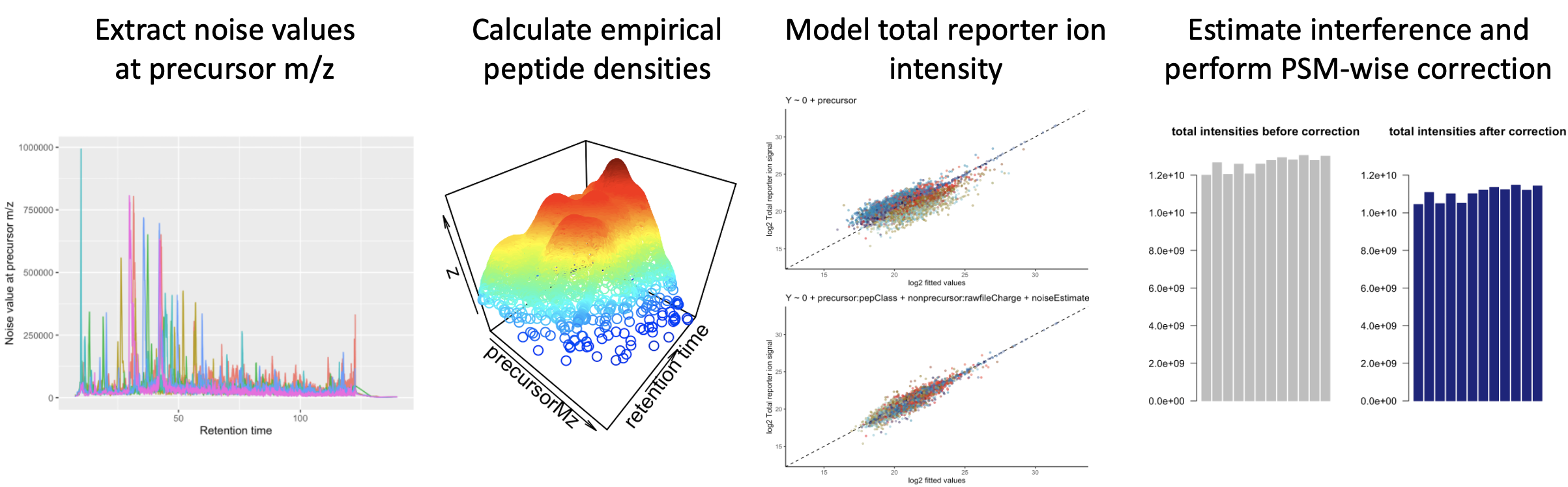
An R implementation for interference modeling and subsequent interference correction in MS2-based multiplex proteomics. Further contains a demo dataset + userguide to get familiar with the workflow. Here are selected visuals from said demo:
This workflow requires the rawStallion Windows command line application to read Thermo raw files and write relevant data to tsv files. Download here.
-
IM.Rmd : R markdown script that performs the entire computational workflow on the basis of specified input parameters.
-
functions_IM.R : Functions required in IM.Rmd script.
-
Demo : A folder containing a demo dataset + userguide. Check out Userguide.pdf contained in this folder for detailed instructions and explanations on the demo and the workflow in general.
-
A PSM table. Currently supported are MaxQuant's msms.txt and Fragpipe's psm.tsv. Other formats might require minor adjustments to the script.
-
Corresponding Thermo raw files used in the database search, located in a separate folder.
-
An isotopic impurity matrix for isotopic impurity correction. Details on the required format are described in the parameter section of IM.Rmd.
- A modified PSM table named modified_PSM.txt. This PSM table contains additional columns such as normalized reporter ion intensities (suffix
_norm), normalized interference-corrected reporter ion intensities (suffix_norm__interference_corrected), as well as several PSM-wise metrics such as Estimated Interference Level (EIL), Precursor Purity Fraction (PPF), and more.
R version 4.1.2 (2021-11-01)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Mojave 10.14.6
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats4 parallel stats graphics grDevices utils datasets methods base
other attached packages:
[1] msqrob2_1.2.0 QFeatures_1.4.0 MultiAssayExperiment_1.20.0 DESeq2_1.34.0
[5] SummarizedExperiment_1.24.0 MatrixGenerics_1.6.0 matrixStats_0.61.0 GenomicRanges_1.46.1
[9] GenomeInfoDb_1.30.0 IRanges_2.28.0 limma_3.50.0 MSnbase_2.20.0
[13] ProtGenerics_1.26.0 S4Vectors_0.32.3 mzR_2.28.0 Rcpp_1.0.7
[17] Biobase_2.54.0 BiocGenerics_0.40.0 cowplot_1.1.1 fields_13.3
[21] viridis_0.6.2 viridisLite_0.4.0 spam_2.8-0 doParallel_1.0.17
[25] iterators_1.0.13 foreach_1.5.2 rlist_0.4.6.2 gridExtra_2.3
[29] MASS_7.3-54 plot3D_1.4 pracma_2.3.8 forcats_0.5.1
[33] stringr_1.4.0 dplyr_1.0.7 purrr_0.3.4 readr_2.1.1
[37] tidyr_1.1.4 tibble_3.1.6 ggplot2_3.3.6 tidyverse_1.3.1
-
Msnbase: Gatto, L. & Lilley, K. S. Msnbase-an R/Bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics 28, 288–289 (2012).
-
fields: Douglas Nychka, Reinhard Furrer, John Paige, S. S. (2021). “fields: Tools for spatial data.”
-
limma: Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
-
DESeq2: Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
-
msqrob2: Goeminne, L. J. E., Gevaert, K. & Clement, L. Peptide-level robust ridge regression improves estimation, sensitivity, and specificity in data-dependent quantitative label-free shotgun proteomics. Mol. Cell. Proteomics 15, 657–668 (2016).
-
MaxQuant: Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
-
plot3D: Soetaert, K. plot3D: Plotting Multi-Dimensional Data.
-
ggplot2: Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag New York).