The Molecular Simulation and Design Framework (MoSDeF) is a collection of open-source tools (hosted on Github) aims at facilitating the construction and simulation of complex molecular systems - with a particular focus on the automated screening of large structural parameter spaces. All tools are written as Python packages and feature a Python-based API.
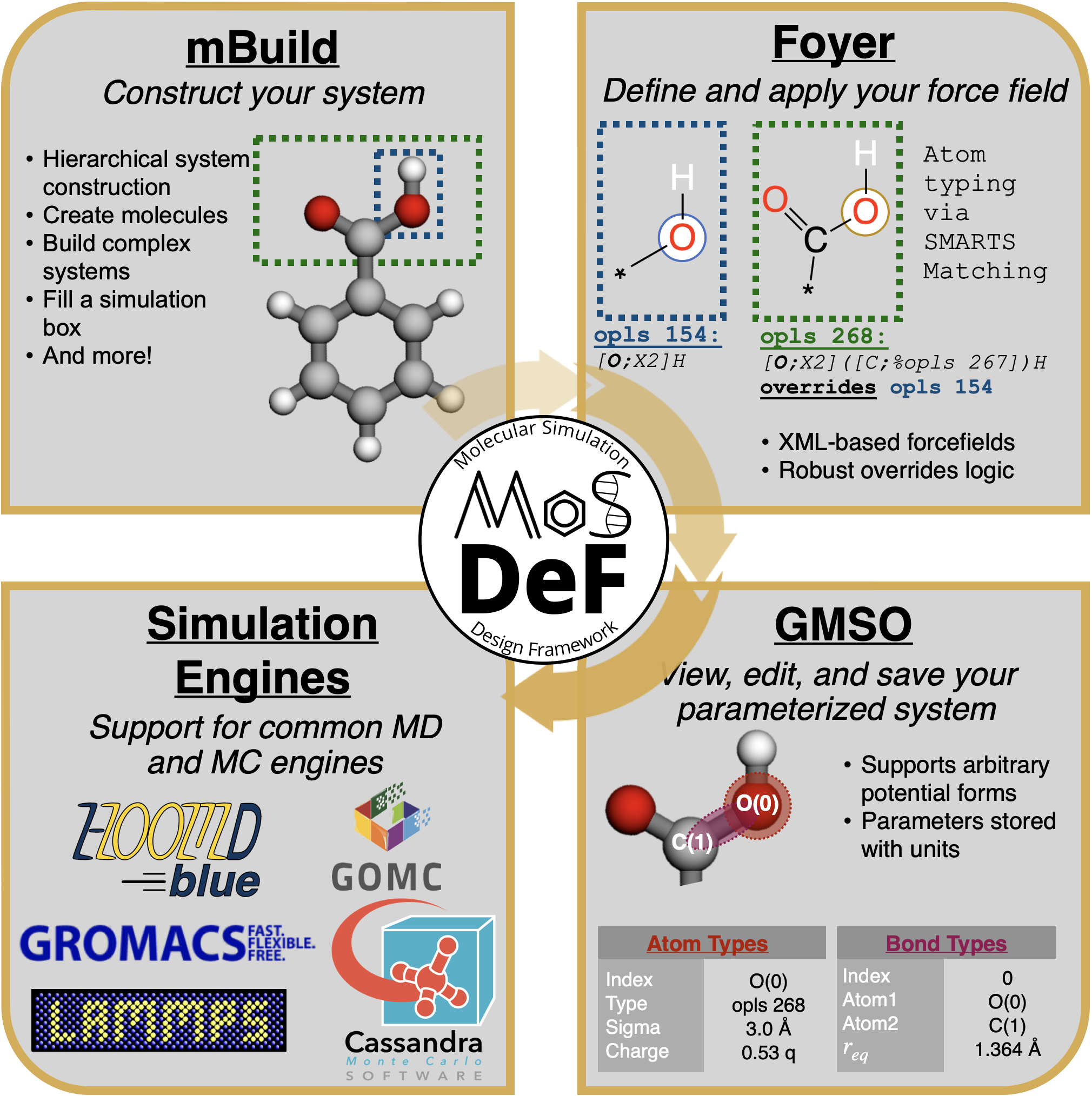
The MoSDeF software suite consist of three core libraries, namely mBuild, Foyer, and GMSO. Each library dedicates to handle a certain step of the chemical system initialization process, as summarized in the figure below.
Libraries in the MoSDeF ecosystem are designed to provide utilities necessary to streamline a researcher's simulation workflow. When setting up simulation studies, we also recommend users to follow the TRUE (Transparent, Reproducible, Usable-by-others, and Extensible) standard, which is a set of common practices meant to improve the reproducibility of computational simulation research.
Due to time constraint, this workshop could not cover all the details and features that offers by the MoSDeF
tools. However, we will visit several key data structures and features that would allow new users to quickly
onboard and start using MoSDeF on their research. Together, we will walk through a set of tutorials, focus on
individual tools in the software suite, namely mBuild, Foyer and GMSO. These tutorials will
be presented in a series of Jupyter notebook, which participants can choose to download and fire up these
notebooks locally or access it through Mybinder.
If you choose to download the tutorials and host them locally, please follow the installation instruction below.
Please leave your comment question during this workshop to the etherpad link: https://etherpad.boisestate.edu/p/mosdef-fomms2022
Follow the command below to clone and create the conda environment for this repository. Assuming you already have conda installed locally:
conda install mamba -n base -c conda-forge
git clone https://github.com/mosdef-hub/fomms-workshop.git
cd fomms-workshop
mamba env create -f environment.yml
mamba activate mosdef-workshop
jupyter notebookFollow the image above to test this repository through the Mybinder service, or follow this link: https://mybinder.org/v2/gh/mosdef-hub/FOMMS-workshop/HEAD
More in-depth MoSDeF tutorials: https://github.com/mosdef-hub/mosdef_tutorials
Example MoSDeF workflow: https://github.com/mosdef-hub/mosdef-workflows
Documentations:
- https://mbuild.mosdef.org/en/stable/
- https://foyer.mosdef.org/en/stable/
- https://gmso.mosdef.org/en/stable/
Join our mailing list: https://groups.google.com/g/mosdef-users