Some tips / things of notes for myself while I'm learning Nextflow
- write notes on the newest nextflow release https://www.nextflow.io/blog/2024/nextflow-2404-highlights.html
Nextflow is a framework used for writing parallel and distributed computational pipelines, common in bioinformatics, genomics, and other fields where complex data analysis pipelines are common. It simplifies the process of creating complex workflows that involve processing large volumes of data.
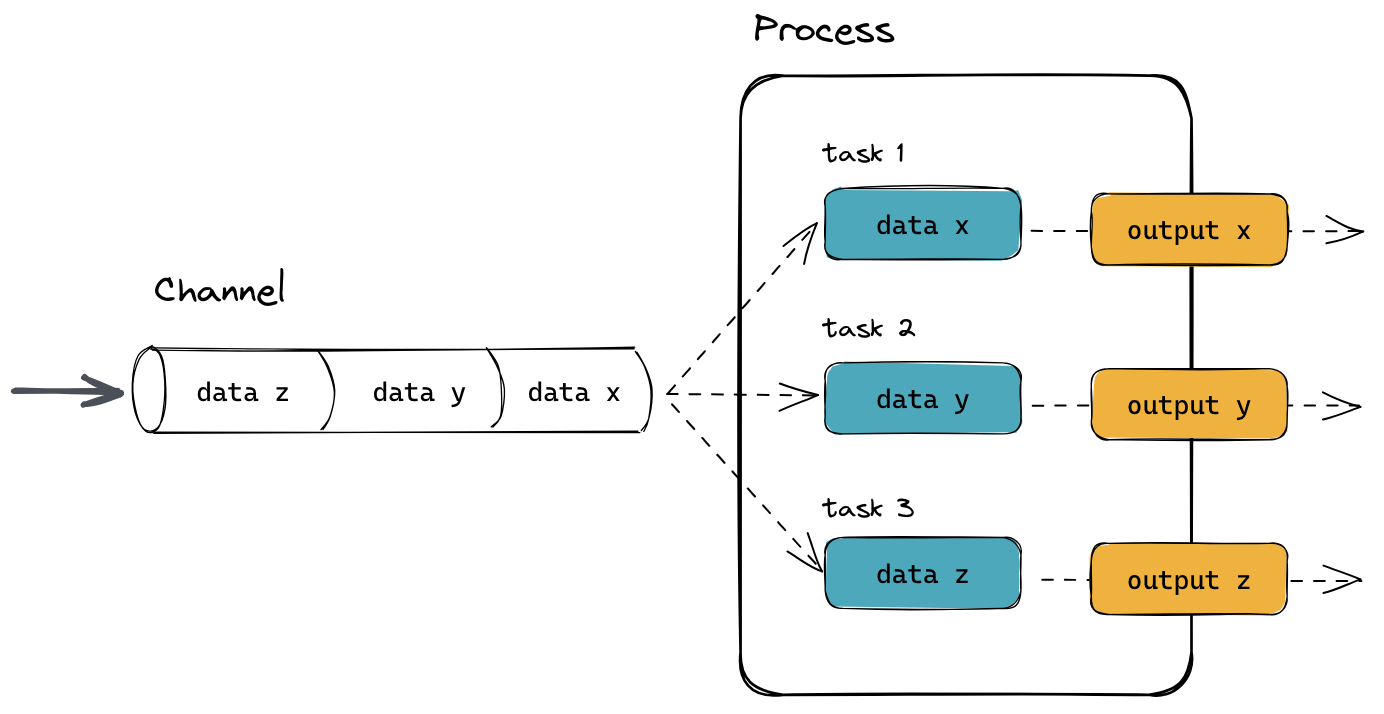
Nextflow pipelines are partitioned into channels, processes, and workflows. Channels represent data streams, processes perform computations on that data, and workflows orchestrate the execution of processes and manage the flow of data through the pipeline.
The central tenant of Nextflow is generating pipelines with 100% reproducibility. Most bioinformatic studies use multiple softwares and languages all with different versions. This makes reproducibility very difficult as you have to go back and figure out what softwares, versions and what dependancies were used. Worst off different softwares might not work on certain machines or even can work differently depending on what machine you are running the analysis on, resulting in different statistical calculations and outputs from the same input data (see https://doi.org/10.1038/nbt.3820 for more information). With Nextflow, if you use it correctly and follow all the best practices, your data analysis pipelines should be fully reproducible regardless of when or where it is run.
Nextflow pipelines need to be written into files with the .nf extension.
Example, create a file called main.nf and provide it the following code:
#!/usr/bin/env nextflow
process FASTQC {
input:
path input
output:
path "*_fastqc.{zip, html}"
script:
"""
fastqc -q $input
"""
}
workflow {
Channel.fromPath("*.fastq.gz") | FASTQC
}Note
The shebang (#!/usr/bin/env nextflow) is a line that helps the operating system decide what program should be used to interpret this script code. If you always use Nextflow to call this script, this line is optional.
Nextflow can then run by using the following command
nextflow run main.nfOnly run it like this the first time. If you run it again it will begin running the entire workflow over again. It's better to not rerun processes that don't need to be rerun to save a lot of time.
When you run a Nextflow pipeline, you will get an output like this in your terminal
N E X T F L O W ~ version 23.10.0
Launching `main.nf` [focused_noether] DSL2 - revision: 197a0e289a
executor > local (3)
[18/f6351b] process > SPLITLETTERS (1) [100%] 1 of 1 ✔
[2f/007bc5] process > CONVERTTOUPPER (1) [100%] 2 of 2 ✔
WORLD!
HELLO
This is what this all means
N E X T F L O W ~
# THE VERSION OF NEXTFLOW THAT WAS USED FOR THIS RUN SPECIFICALLY
version 23.10.0
# THE NAME OF THE SCRIPT FILE THAT WAS RUN WITH NEXTFLOW
Launching `main.nf`
# A GENERATED MNEMONIC WHICH IS AN RANDOM OBJECTIVE AND A RANDOM SURNAME OF A FAMOUS SCIENTIST
[focused_noether]
# THE VERSION OF THE NEXTFLOW LANGUAGE
DSL2
# THE REVISION HASH, WHICH IS LIKE AN ID OF YOUR PIPELINE.
# IF YOU CHANGE THE NEXTFLOW SCRIPT CODE, THIS REVISION ID WILL CHANGE
- revision: 197a0e289a
# THE EXECUTOR FOR THE PIPELINE
executor > local
# NEXTFLOW'S GUESS AT HOW MANY TASKS THAT WILL OCCUR IN YOUR PIPELINE
(3)
# A LIST OF PROCESSES AND TASKS
[18/f6351b] process > SPLITLETTERS (1) [100%] 1 of 1 ✔
[2f/007bc5] process > CONVERTTOUPPER (1) [100%] 2 of 2 ✔
## EVERY TASK WILL HAVE A UNIQUE HASH
## EVERY TASK IS ISOLATED FROM EACH OTHER
## THESE HASHES CORRESPOND TO DIRECTORY NAMES WHERE YOU CAN GO AND VIEW INFORMATION ABOUT THAT SPECIFIC TASK
[18/f6351b]
[2f/007bc5] # BY DEFAULT NEXTFLOW SHOWS THE HASH OF 1 TASK PER PROCESS.
# THE WORKFLOW OUTPUTS
WORLD!
HELLO
When using the -resume flag, successfully completed tasks are skipped and the previously cached results are used in downstream tasks.
nextflow run main.nf -resumeIn practice, every execution starts from the beginning. However, when using resume, before launching a task, Nextflow uses the unique ID to check if:
- the working directory exists
- it contains a valid command exit status
- it contains the expected output files.
The mechanism works by assigning a unique ID to each task. This unique ID is used to create a separate execution directory, called the working directory, where the tasks are executed and the results stored. A task’s unique ID is generated as a 128-bit hash number obtained from a composition of the task’s:
- Inputs values
- Input files
- Command line string
- Container ID
- Conda environment
- Environment modules
- Any executed scripts in the bin directory
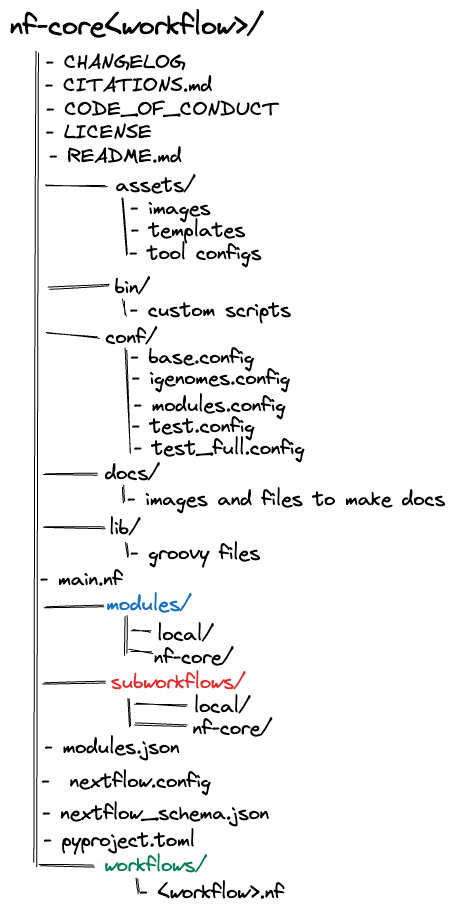
An basic, overall professional Nextflow pipeline structure should look like this:
.
├── main.nf
├── modules
│ ├── local
│ └── nf-core
├── subworkflows
│ ├── local
│ └── nf-core
└── workflows
├── illumina.nf
├── nanopore.nf
└── sra_download.nf
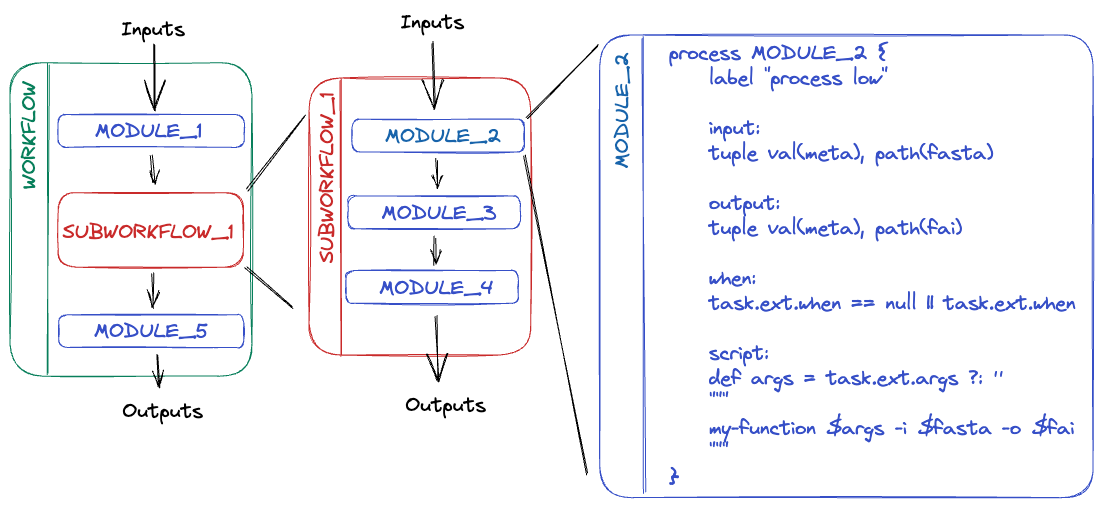
Terminology:
- Module:
- An atomic process
- e.g. a module containing a single tool such as FastQC
- An atomic process
- Subworkflow:
- A chain of multiple modules with a higher-level of functionality
- e.g. a subworkflow to run multiple QC tools on FastQ files
- A chain of multiple modules with a higher-level of functionality
- Workflows:
- An end-to-end pipeline
- DSL1: A large monolithic script
- DSL2: A combination of individual modules and subworkflows
- e.g. a pipeline to produce a series of final outputs from one or more inputs.
main.nf:- The entry point for Nextflow
- Where all the workflows will be called from
Note
DSL1 and DSL2 refer to two versions of the Nextflow Domain Specific Language (DSL).
The first thing that Nextflow looks for when a workflow is run is configuration files in multiple locations. Since each configuration file can contain conflicting settings, the sources are ranked to determine which settings are applied. Possible configuration sources, in order of priority:
-
Parameters specified on the command line (
--something value) -
Parameters provided using the
-params-fileoption -
Config file specified using the
-c my_configoption -
The config file named
nextflow.configin the current directory -
The config file named
nextflow.configin the workflow project directory -
The config file
$HOME/.nextflow/config -
Values defined within the pipeline script itself (e.g.
main.nf)
Note
Precedence is in order of 'distance'. A handy guide to understand configuration precedence is in order of 'distance from the command-line invocation'. Parameters specified directly on the CLI --example foo are "closer" to the run than configuration specified in the remote repository.
When more than one of these options for specifying configurations are used, they are merged, so that the settings in the first override the same settings appearing in the second, and so on.
This is an example of a Nextflow configuration in a file called nextflow.config
propertyOne = 'world'
propertyTwo = "Hello $propertyOne"
customPath = "$PATH:/my/app/folder"Note
The quotes act like they do in bash. Variables inside single quotes remain literal. Variables inside double quotes get expanded (including environment variables)
Configuration settings can be organized in different scopes by dot prefixing the property names with a scope identifier, or grouping the properties in the same scope using the curly brackets notation. For example:
alpha.x = 1
alpha.y = 'string value..'
beta {
p = 2
q = 'another string ..'
}The scope params allows the definition of workflow parameters that override the values defined in the main workflow script.
This is useful to consolidate one or more execution parameters in a separate file.
nextflow.config
params.foo = 'Bonjour'
params.bar = 'le monde!'snippet.nf
params.foo = 'Hello'
params.bar = 'world!'
// print both params
println "$params.foo $params.bar"Any variables that are set using params. can be modified through the command line when executing Nextflow using two dashes (--)
E.g.
nextflow run snippet.nf -resume --foo "Bye"Note
In Nextflow, single dashes (-) in command line arguments refer to Nextflow commands (e.g. -resume), while double dashes (--) refer to workflow parameters
Note
Values assigned to a the config params params. will be treated as value channels (see more information below on value channels)
The env scope allows the definition of one or more variables that will be exported into the environment where the workflow tasks will be executed.
env.NCBI_SETTINGS = params.settings_fileProcess directives allow the specification of settings for the task execution such as cpus, memory, container, and other resources in the workflow script.
This is useful when prototyping a small workflow script.
However, it’s always a good practice to decouple the workflow execution logic from the process configuration settings, i.e. it’s strongly suggested to define the process settings in the workflow configuration file instead of the workflow script.
The process configuration scope allows the setting of any process directives in the Nextflow configuration file:
nextflow.config
process {
cpus = 10
memory = 8.GB
container = 'biocontainers/bamtools:v2.4.0_cv3'
}The above config snippet defines the cpus, memory and container directives for all processes in your workflow script. Depending on your executor these things may behave differently.
The withLabel selectors allow the configuration of all processes annotated with a label directive as shown below:
process {
withLabel: big_mem {
cpus = 16
memory = 64.GB
queue = 'long'
}
}The above configuration example assigns 16 cpus, 64 Gb of memory and the long queue to all processes annotated with the big_mem label.
Labels can be added using the label directive
e.g.
process bigTask {
label 'big_mem'
```
<task script>
```
}In the same manner, the withName selector allows the configuration of a specific process in your pipeline by its name. For example:
process {
withName: hello {
cpus = 4
memory = 8.GB
queue = 'short'
}
}A process selector can also be negated prefixing it with the special character !. For example:
process {
withLabel: 'foo' { cpus = 2 }
withLabel: '!foo' { cpus = 4 }
withName: '!align.*' { queue = 'long' }
}The above configuration snippet sets 2 cpus for the processes annotated with the foo label and 4 cpus to all processes not annotated with that label. Finally it sets the use of long queue to all process whose name does not start with align.
Both label and process name selectors allow the use of a regular expression in order to apply the same configuration to all processes matching the specified pattern condition.
For example:
process {
withLabel: 'foo|bar' {
cpus = 2
memory = 4.GB
}
}The above configuration snippet sets 2 cpus and 4 GB of memory to the processes annotated with a label foo and bar.
A process selector can be negated prefixing it with the special character !.
For example:
process {
withLabel: 'foo' { cpus = 2 }
withLabel: '!foo' { cpus = 4 }
withName: '!align.*' { queue = 'long' }
}The above configuration snippet sets 2 cpus for the processes annotated with the foo label and 4 cpus to all processes not annotated with that label. Finally it sets the use of long queue to all process whose name does not start with align.
If you already have a conda environment in your machine that you want to use for your processes, you can use
nextflow.config
process.conda = "/home/ubuntu/miniconda2/envs/nf-tutorial"
You can specify the path of an existing Conda environment as either directory or the path of Conda environment YAML file.
When a workflow script is launched, Nextflow looks for a file named nextflow.config in the current directory and in the script base directory (if it is not the same as the current directory). Finally, it checks for the file: $HOME/.nextflow/config.
When more than one of the above files exists, they are merged, so that the settings in the first override the same settings that may appear in the second, and so on.
The default config file search mechanism can be extended by providing an extra configuration file by using the command line option: -c <config file>.
Information on writing these config files can be found here https://training.nextflow.io/basic_training/config/.
If you add the following code to the nextflow.config file
process.executor = 'slurm'Then Nextflow will write the SLURM job script for every file for you. Nextflow will manage each process as a separate job that is submitted to the cluster using the sbatch command.
More information on how to configure this further can be found here https://www.nextflow.io/docs/latest/executor.html#slurm
When running Nextflow on a HPC, it's recommended to run it as a job on a compute node. This is because a lot of computing clusters have strict rules on running processes on login nodes. Therefore, it's always advisable to create jobscripts like this for all your Nextflow jobs.
launch_nf.sh
#!/bin/bash
#SBATCH --partition WORK
#SBATCH --mem 5G
#SBATCH -c 1
#SBATCH -t 12:00:00
WORKFLOW=$1
CONFIG=$2
# Use a conda environment where you have installed Nextflow
# (may not be needed if you have installed it in a different way)
conda activate Nextflow
nextflow -C ${CONFIG} run ${WORKFLOW}and launch the workflow using
sbatch launch_nf.sh /home/my_user/path/my_workflow.nf /home/my_user/path/my_config_file.confThe following information about configuration is taken from the Nextflow advanced training.
There may be some configuration values that you will want applied on all runs for a given system. These configuration values should be written to ~/.nextflow/config.
For example - you may have an account on a HPC system and you know that you will always want to submit jobs using the SLURM scheduler when using that machine and always use the Singularity container engine. In this case, your ~/.nextflow/config file may include:
process.executor = 'slurm'
singularity.enable = trueThese configuration values would be inherited by every run on that system without you needing to remember to specify them each time.
Process directives (listed here) can be overridden using the process block.
For example, if we wanted to specify that all tasks for a given run should use 2 cpus. In the nextflow.config file in the current working directory add:
process {
cpus = 2
}... and then run:
nextflow run rnaseq-nfWe can make the configuration more specific by using process selectors.
We can use process names and/or labels to apply process-level directives to specific tasks:
process {
withName: 'RNASEQ:INDEX' {
cpus = 2
}
}Glob pattern matching can also be used:
process {
withLabel: '.*:INDEX' {
cpus = 2
}
}This will run any task that ends with :INDEX to run with 2 cpus.
Just to make sure the globbing is working, you can also configure your processes to have a tag directive. The process tag is the value that's printed out in parentheses after the process name.
For example, when run the Nextflow pipeline with the above configuration will output this
N E X T F L O W ~ version 23.04.3
Launching `https://github.com/nextflow-io/rnaseq-nf` [fabulous_bartik] DSL2 - revision: d910312506 [master]
R N A S E Q - N F P I P E L I N E
===================================
transcriptome: /home/gitpod/.nextflow/assets/nextflow-io/rnaseq-nf/data/ggal/ggal_1_48850000_49020000.Ggal71.500bpflank.fa
reads : /home/gitpod/.nextflow/assets/nextflow-io/rnaseq-nf/data/ggal/ggal_gut_{1,2}.fq
outdir : results
executor > local (4)
[1d/3c5cfc] process > RNASEQ:INDEX (ggal_1_48850000_49020000) [100%] 1 of 1 ✔
[38/a6b717] process > RNASEQ:FASTQC (FASTQC on ggal_gut) [100%] 1 of 1 ✔
[39/5f1cc4] process > RNASEQ:QUANT (ggal_gut) [100%] 1 of 1 ✔
[f4/351d02] process > MULTIQC [100%] 1 of 1 ✔
Done! Open the following report in your browser --> results/multiqc_report.html
If you modify the nextflow.config to look like this:
process {
withLabel: '.*:INDEX' {
cpus = 2
tag = "Example debug"
}
}The output will then turn into this, showing that only the processes with :INDEX will be affected.
N E X T F L O W ~ version 23.04.3
Launching `https://github.com/nextflow-io/rnaseq-nf` [fabulous_bartik] DSL2 - revision: d910312506 [master]
R N A S E Q - N F P I P E L I N E
===================================
transcriptome: /home/gitpod/.nextflow/assets/nextflow-io/rnaseq-nf/data/ggal/ggal_1_48850000_49020000.Ggal71.500bpflank.fa
reads : /home/gitpod/.nextflow/assets/nextflow-io/rnaseq-nf/data/ggal/ggal_gut_{1,2}.fq
outdir : results
executor > local (4)
[1d/3c5cfc] process > RNASEQ:INDEX (Example debug) [100%] 1 of 1 ✔
[38/a6b717] process > RNASEQ:FASTQC (FASTQC on ggal_gut) [100%] 1 of 1 ✔
[39/5f1cc4] process > RNASEQ:QUANT (ggal_gut) [100%] 1 of 1 ✔
[f4/351d02] process > MULTIQC [100%] 1 of 1 ✔
Done! Open the following report in your browser --> results/multiqc_report.html
We can specify dynamic directives using closures that are computed as the task is submitted. This allows us to (for example) scale the number of CPUs used by a task by the number of input files. This is one of the benefits of Nextflow, which is that because it is such a dynamic paradigm, there are elements of the task that can be calculated as the task is being executed, rather than before when the run begins.
Give the FASTQC process in the rnaseq-nf workflow
process FASTQC {
tag "FASTQC on $sample_id"
conda 'fastqc=0.12.1'
publishDir params.outdir, mode:'copy'
input:
tuple val(sample_id), path(reads)
output:
path "fastqc_${sample_id}_logs"
script:
"""
fastqc.sh "$sample_id" "$reads"
"""
}We might choose to scale the number of CPUs for the process by the number of files in reads:
process {
withName: 'FASTQC' {
cpus = { reads.size() }
}
}Remember, dynamic directives are supplied a closure, in this case { reads.size() }. This is telling Nextflow "I want to defer evaluation of this. I'm just supplying a closure that will be evaluated later" (Think like callback functions in JavaScript).
We can even use the size of the input files. Here we simply sum together the file sizes (in bytes) and use it in the tag block:
process {
withName: 'FASTQC' {
cpus = { reads.size() }
tag = {
total_bytes = reads*.size().sum()
"Total size: ${total_bytes as MemoryUnit}"
}
}
}Note
MemoryUnit is a Nextflow built-in class for turning strings and integers into human-readable memory units.
When we run this:
N E X T F L O W ~ version 23.04.3
Launching `https://github.com/nextflow-io/rnaseq-nf` [fabulous_bartik] DSL2 - revision: d910312506 [master]
R N A S E Q - N F P I P E L I N E
===================================
transcriptome: /home/gitpod/.nextflow/assets/nextflow-io/rnaseq-nf/data/ggal/ggal_1_48850000_49020000.Ggal71.500bpflank.fa
reads : /home/gitpod/.nextflow/assets/nextflow-io/rnaseq-nf/data/ggal/ggal_gut_{1,2}.fq
outdir : results
executor > local (4)
[1d/3c5cfc] process > RNASEQ:INDEX (ggal_1_48850000_49020000) [100%] 1 of 1 ✔
[38/a6b717] process > RNASEQ:FASTQC (Total size: 1.3 MB) [100%] 1 of 1 ✔
[39/5f1cc4] process > RNASEQ:QUANT (ggal_gut) [100%] 1 of 1 ✔
[f4/351d02] process > MULTIQC [100%] 1 of 1 ✔
Done! Open the following report in your browser --> results/multiqc_report.html
Note
Dynamic directives need to be supplied as closures encases in curly braces.
Nextflow gives you the option of resubmitting a task if a task fails. This is really useful if you're working with a flakey program or dealing with network I/O.
The most common use for dynamic process directives is to enable tasks that fail due to insufficient memory to be resubmitted for a second attempt with more memory.
To enable this, two directives are needed:
maxRetrieserrorStrategy
The errorStrategy directive determines what action Nextflow should take in the event of a task failure (a non-zero exit code). The available options are:
terminate: (default) Nextflow terminates the execution as soon as an error condition is reported. Pending jobs are killedfinish: Initiates an orderly pipeline shutdown when an error condition is raised, waiting the completion of any submitted job.ignore: Ignores processes execution errors. (dangerous)retry: Re-submit for execution a process returning an error condition.
If the errorStrategy is retry, then it will retry up to the value of maxRetries times.
If using a closure to specify a directive in configuration, you have access to the task variable, which includes the task.attempt value - an integer specifying how many times the task has been retried. We can use this to dynamically set values such as memory and cpus
process {
withName: 'RNASEQ:QUANT' {
errorStrategy = 'retry'
maxRetries = 3
memory = { 2.GB * task.attempt }
time = { 1.hour * task.attempt }
}
}Note
Be aware of the differences between configuration and process syntax differences.
- When defining values inside configuration, an equals sign
=is required as shown above. - When specifying process directives inside the process (in a
.nffile), no=is required:
process MULTIQC {
cpus 2
// ...In Nextflow, a process is the basic computing primitive to execute foreign functions (i.e., custom scripts or tools).
The process definition starts with the keyword process, followed by the process name and finally the process body delimited by curly brackets.
It is a best practice to always name processes in UPPERCASE. This way you can easily see what are process blocks and what are regular functions.
A basic process, only using the script definition block, looks like the following:
process SAYHELLO {
script:
"""
echo 'Hello world!'
"""
}However, the process body can contain up to five definition blocks:
-
Directives are initial declarations that define optional settings
-
Input defines the expected input channel(s)
- Requires a qualifier
-
Output defines the expected output channel(s)
- Requires a qualifier
-
When is an optional clause statement to allow conditional processes
-
Script is a string statement that defines the command to be executed by the process' task
The full process syntax is defined as follows:
process < name > {
[ directives ]
input:
< process inputs >
output:
< process outputs >
when:
< condition >
[script|shell|exec]:
"""
< user script to be executed >
"""
}Take a look at the last part of the output when you run a Nextflow pipeline
# YOUR PROCESSES AND OUTPUTS
[18/f6351b] process > SPLITLETTERS (1) [100%] 1 of 1 ✔
[2f/007bc5] process > CONVERTTOUPPER (1) [100%] 2 of 2 ✔
WORLD!
HELLO
Breaking down the processes section further this is what each part means
# THE HEXADECIMAL HASH FOR THE TASK (ALSO THE TASK DIRECTORY NAME)
[18/f6351b]
# THE NAME OF THE PROCESS USED FOR THE TASK
process > SPLITLETTERS
# I'M NOT SURE WHAT THIS NUMBER MEANS YET. MAYBE THE CURRENT PROCESS?
(1)
# THE PROGRESS OF ALL THE TASKS (OR MAYBE THE CURRENT TASK?)
[100%]
# ENUMERATED PROGRESS OF ALL THE TASKS
1 of 1
# SIGN SHOWING THE TASK RAN SUCCESSFULLY
✔
# THE HEXADECIMAL HASH FOR THE TASK (LAST TASK ONLY IF THERE ARE MORE THAN ONE)
[2f/007bc5]
# THE NAME OF THE PROCESS USED FOR THE TASK
process > CONVERTTOUPPER
# I'M NOT SURE WHAT THIS NUMBER MEANS YET. MAYBE THE CURRENT PROCESS?
(1)
# THE PROGRESS OF ALL THE TASKS (OR MAYBE THE CURRENT TASK?)
[100%]
# ENUMERATED PROGRESS OF ALL THE TASKS
2 of 2
# SIGN SHOWING THE TASK RAN SUCCESSFULLY
✔
The hexadecimal numbers, like 18/f6351b, identify the unique process execution, that is called a task. These numbers are also the prefix of the directories where each task is executed. You can inspect the files produced by changing to the directory $PWD/work and using these numbers to find the task-specific execution path (e.g. Go to $PWD/work/18/f6351b46bb9f65521ea61baaaa9eff to find all the information on the task performed using the SPLITLETTERS process).
Note
Inside the work directory for the specific task, you will also find the Symbolic links used as inputs for the specific task, not copies of the files themselves.
If you look at the second process in the above examples, you notice that it runs twice (once for each task), executing in two different work directories for each input file. The ANSI log output from Nextflow dynamically refreshes as the workflow runs; in the previous example the work directory 2f/007bc5 is the second of the two directories that were processed (overwriting the log with the first). To print all the relevant paths to the screen, disable the ANSI log output using the -ansi-log flag.
Example
nextflow run hello.nf -ansi-log falseWill output
N E X T F L O W ~ version 23.10.0
Launching `hello.nf` [boring_bhabha] DSL2 - revision: 197a0e289a
[18/f6351b] Submitted process > SPLITLETTERS (1)
[dc/e177f3] Submitted process > CONVERTTOUPPER (1) # NOTICE THE HASH FOR TASK 1 IS VISIBLE NOW
[2f/007bc5] Submitted process > CONVERTTOUPPER (2)
HELLO
WORLD!
Now you can find out in which directory everything related to every task performed is stored straight from the console.
Note
Even if you don't use the -ansi-log false flag, you can still see the hashes/directories all the tasks are stored in using the .nextflow.log file. The task directories can be found in the [Task monitor] logs.
Every task that is executed by Nextflow will produe a bunch of hidden files in the tasks work directory beginning with .command. Below are a list of all of them and what they contain.
This file is created whenever the task really started.
Whenever you are debugging a pipeline and you don't know if a task really started or not, you can check for the existence of this file.
This file contains all the errors that may have occured for this task.
This file contains the logs created for this task (e.g. with log.info or through other methods).
This file contains anything that was printed to your screen (the standard output).
This file shows you the jobscript that Nextflow created to run the script (e.g. If you are running your scripts with SLURM, it will show you the SLURM job script Nextflow created and that was subsequently called with sbatch).
This script contains all the functions Nextflow needs to make sure your script runs on whatever executor you configured (e.g. locally, in the cloud, on a HPC, with or withouth container, etc.)
You're not really supposed to meddle with this file but sometimes you may want to see what's in it. (E.g. To see what Docker command was used to start the container etc.)
This file contains the final script that was run for that task.
Example: If this is in the workflow
params.greeting = 'Hello world!'
process SPLITLETTERS {
input:
val x
output:
path 'chunk_*'
"""
printf '$x' | split -b 6 - chunk_
"""
}
workflow {
letters_ch = SPLITLETTERS(greeting_ch)
}The .command.sh file for a task run on this process will look like this
printf 'Hello world!' | split -b 6 - chunk_This is very useful for troubleshooting when things don't work like you'd expect.
The log.info command can be used to print multiline information using groovy’s logger functionality. Instead of writing a series of println commands, it can be used to include a multiline message.
log.info """\
R N A S E Q - N F P I P E L I N E
===================================
transcriptome: ${params.transcriptome_file}
reads : ${params.reads}
outdir : ${params.outdir}
"""
.stripIndent(true)log.info not only prints the ouput of the command to the screen (stout), but also prints the results to the log file.
Usually when you write Nextflow scripts you will add indenting so you can better read the code. However when you want to print the code to the screen you often don't want indenting. The .stripIndent(true) method removes the indents from the output.
.view is a channel operator that consumes every element of a channel and prints it to the screen.
Example
// main.nf
params.reads = "${baseDir}/data/reads/ENCSR000COQ1_{1,2}.fastq.gz"
workflow {
reads_ch = Channel.fromFilePairs(params.reads)
reads_ch.view()
}nextflow run main.nf
# Outputs
# [ENCSR000COQ1, [path/to/ENCSR000COQ1_1.fastq.gz, path/to/ENCSR000COQ1_2.fastq.gz]]You can see in this case it outputs a single channel element created from the .fromFilePairs channel operator.
- The first item is an ID (in this case the replicate ID)
- The second item is a list with all the items (in this case the path to the reads)
In addition to the argument-less usage of view as shown above, this operator can also take a closure to customize the stdout message. We can create a closure to print the value of the elements in a channel as well as their type, for example.
def timesN = { multiplier, it -> it * multiplier }
def timesTen = timesN.curry(10)
workflow {
Channel.of( 1, 2, 3, 4, 5 )
| map( timesTen )
| view { "Found '$it' (${it.getClass()})"}
}The input block allows you to define the input channels of a process, similar to function arguments. A process may have at most one input block, and it must contain at least one input.
The input block follows the syntax shown below:
input:
<input qualifier> <input name>An input definition consists of a qualifier and a name. The input qualifier defines the type of data to be received. This information is used by Nextflow to apply the semantic rules associated with each qualifier, and handle it properly depending on the target execution platform (grid, cloud, etc).
The following input qualifiers are available:
val: Access the input value by name in the process script.
file: (DEPRECATED) Handle the input value as a file, staging it properly in the execution context.
path: Handle the input value as a path, staging the file properly in the execution context.
env: Use the input value to set an environment variable in the process script.
stdin: Forward the input value to the process stdin special file.
tuple: Handle a group of input values having any of the above qualifiers.
each: Execute the process for each element in the input collection.
More information on how each of these input qualifiers work can be found here https://www.nextflow.io/docs/latest/process.html#inputs
A key feature of processes is the ability to handle inputs from multiple channels. However, it’s important to understand how channel contents and their semantics affect the execution of a process.
Consider the following example:
ch1 = Channel.of(1, 2, 3)
ch2 = Channel.of('a', 'b', 'c')
process FOO {
debug true
input:
val x
val y
script:
"""
echo $x and $y
"""
}
workflow {
FOO(ch1, ch2)
}Ouputs:
1 and a
3 and c
2 and b
Both channels emit three values, therefore the process is executed three times, each time with a different pair.
The process waits until there’s a complete input configuration, i.e., it receives an input value from all the channels declared as input.
When this condition is verified, it consumes the input values coming from the respective channels, spawns a task execution, then repeats the same logic until one or more channels have no more content.
This means channel values are consumed serially one after another and the first empty channel causes the process execution to stop, even if there are other values in other channels.
What happens when channels do not have the same cardinality (i.e., they emit a different number of elements)?
ch1 = Channel.of(1, 2, 3)
ch2 = Channel.of('a')
process FOO {
debug true
input:
val x
val y
script:
"""
echo $x and $y
"""
}
workflow {
FOO(ch1, ch2)
}Outputs:
1 and a
In the above example, the process is only executed once because the process stops when a channel has no more data to be processed.
However, replacing ch2 with a value channel will cause the process to be executed three times, each time with the same value of a.
For more information see here.
The each qualifier allows you to repeat the execution of a process for each item in a collection every time new data is received.
This is a very good way to try multiple parameters for a certain process.
Example:
sequences = Channel.fromPath("$baseDir/data/ggal/*_1.fq")
methods = ['regular', 'espresso']
process ALIGNSEQUENCES {
debug true
input:
path seq
each mode
script:
"""
echo t_coffee -in $seq -mode $mode
"""
}
workflow {
ALIGNSEQUENCES(sequences, methods)
}Outputs:
t_coffee -in gut_1.fq -mode regular
t_coffee -in lung_1.fq -mode espresso
t_coffee -in liver_1.fq -mode regular
t_coffee -in gut_1.fq -mode espresso
t_coffee -in lung_1.fq -mode regular
t_coffee -in liver_1.fq -mode espresso
In the above example, every time a file of sequences is received as an input by the process, it executes three tasks, each running a different alignment method set as a mode variable. This is useful when you need to repeat the same task for a given set of parameters.
A double asterisk (**) in a glob pattern works like * but also searches through subdirectories. For example, imagine this is your file structure
data
├── 2023-11-08_upn22_rep-1_mhmw--100mg
│ └── no_sample
│ └── 20231108_1310_MC-115499_FAX00407_d135d0ec
│ ├── fastq_fail
│ │ ├── FAX00407_fail_d135d0ec_b0bb43ca_0.fastq.gz
│ │ └── FAX00407_fail_d135d0ec_b0bb43ca_1.fastq.gz
│ ├── fastq_pass
│ │ ├── FAX00407_pass_d135d0ec_b0bb43ca_0.fastq.gz
│ │ └── FAX00407_pass_d135d0ec_b0bb43ca_1.fastq.gz
│ ├── other_reports
│ │ ├── pore_scan_data_FAX00407_d135d0ec_b0bb43ca.csv
│ │ └── temperature_adjust_data_FAX00407_d135d0ec_b0bb43ca.csv
│ ├── pod5_fail
│ │ ├── FAX00407_fail_d135d0ec_b0bb43ca_0.pod5
│ │ └── FAX00407_fail_d135d0ec_b0bb43ca_1.pod5
│ └── pod5_pass
│ ├── FAX00407_pass_d135d0ec_b0bb43ca_0.pod5
│ └── FAX00407_pass_d135d0ec_b0bb43ca_1.pod5
├── 2023-11-12_upn22_rep-2_mhmw--recovery-elude-1
│ └── no_sample
│ └── 20231112_1338_MC-115499_FAX00228_b67d08a5
│ ├── fastq_fail
│ │ ├── FAX00228_fail_b67d08a5_dc19481f_0.fastq.gz
│ │ └── FAX00228_fail_b67d08a5_dc19481f_1.fastq.gz
│ ├── fastq_pass
│ │ ├── FAX00228_pass_b67d08a5_dc19481f_0.fastq.gz
│ │ └── FAX00228_pass_b67d08a5_dc19481f_1.fastq.gz
│ ├── final_summary_FAX00228_b67d08a5_dc19481f.txt
│ ├── other_reports
│ │ ├── pore_scan_data_FAX00228_b67d08a5_dc19481f.csv
│ │ └── temperature_adjust_data_FAX00228_b67d08a5_dc19481f.csv
│ ├── pod5_fail
│ │ ├── FAX00228_fail_b67d08a5_dc19481f_0.pod5
│ │ └── FAX00228_fail_b67d08a5_dc19481f_1.pod5
│ ├── pod5_pass
│ │ ├── FAX00228_pass_b67d08a5_dc19481f_0.pod5
│ │ └── FAX00228_pass_b67d08a5_dc19481f_1.pod5
│ └── sequencing_summary_FAX00228_b67d08a5_dc19481f.txt
├── 2023-11-16_upn22_rep-3_mhmw
│ └── no_sample
│ └── 20231116_0945_MC-115499_FAX00393_849b7392
│ ├── barcode_alignment_FAX00393_849b7392_a554d814.tsv
│ ├── fastq_fail
│ │ ├── FAX00393_fail_849b7392_a554d814_0.fastq.gz
│ │ └── FAX00393_fail_849b7392_a554d814_1.fastq.gz
│ ├── fastq_pass
│ │ ├── FAX00393_pass_849b7392_a554d814_0.fastq.gz
│ │ └── FAX00393_pass_849b7392_a554d814_1.fastq.gz
│ ├── final_summary_FAX00393_849b7392_a554d814.txt
│ ├── other_reports
│ │ ├── pore_scan_data_FAX00393_849b7392_a554d814.csv
│ │ └── temperature_adjust_data_FAX00393_849b7392_a554d814.csv
│ ├── pod5_fail
│ │ ├── FAX00393_fail_849b7392_a554d814_0.pod5
│ │ └── FAX00393_fail_849b7392_a554d814_1.pod5
│ ├── pod5_pass
│ │ ├── FAX00393_pass_849b7392_a554d814_0.pod5
│ │ └── FAX00393_pass_849b7392_a554d814_1.pod5
│ ├── pore_activity_FAX00393_849b7392_a554d814.csv
│ ├── report_FAX00393_20231116_0945_849b7392.html
│ ├── report_FAX00393_20231116_0945_849b7392.json
│ ├── report_FAX00393_20231116_0945_849b7392.md
│ ├── sample_sheet_FAX00393_20231116_0945_849b7392.csv
│ ├── sequencing_summary_FAX00393_849b7392_a554d814.txt
│ └── throughput_FAX00393_849b7392_a554d814.csv
└── 2023-10-26_upn22_rep-3_pci
└── no_sample
└── 20231026_1515_MC-115499_FAW96674_9d505d15
├── barcode_alignment__9d505d15_1f674c3a.tsv
├── fastq_fail
│ ├── FAW96674_fail_9d505d15_1f674c3a_0.fastq.gz
│ └── FAW96674_fail_9d505d15_1f674c3a_1.fastq.gz
├── fastq_pass
│ ├── FAW96674_pass_9d505d15_1f674c3a_0.fastq.gz
│ └── FAW96674_pass_9d505d15_1f674c3a_1.fastq.gz
├── final_summary_FAW96674_9d505d15_1f674c3a.txt
├── other_reports
│ ├── pore_scan_data_FAW96674_9d505d15_1f674c3a.csv
│ └── temperature_adjust_data_FAW96674_9d505d15_1f674c3a.csv
├── pod5_fail
│ ├── FAW96674_fail_9d505d15_1f674c3a_0.pod5
│ └── FAW96674_fail_9d505d15_1f674c3a_1.pod5
├── pod5_pass
│ ├── FAW96674_pass_9d505d15_1f674c3a_0.pod5
│ └── FAW96674_pass_9d505d15_1f674c3a_1.pod5
├── pore_activity__9d505d15_1f674c3a.csv
├── report__20231026_1515_9d505d15.html
├── report__20231026_1515_9d505d15.json
├── report__20231026_1515_9d505d15.md
├── sample_sheet__20231026_1515_9d505d15.csv
├── sequencing_summary_FAW96674_9d505d15_1f674c3a.txt
└── throughput__9d505d15_1f674c3a.csv
33 directories, 60 files
All the .fastq.gz files can be grabbed using the ** wildcard to search through all the subdirectories to look for the files with the .fastq extensions.
fastq_ch = Channel.fromPath("data/**/*.fastq.gz").collect()
When you run a program, theres a very high likelihood that many output or intermediate files will be created. what the output: syntax specifies is the only file or files (or stdout) that your want to include in your output channel for the next process or processes.
The output declaration block defines the channels used by the process to send out the results produced.
Only one output block, that can contain one or more output declaration, can be defined. The output block follows the syntax shown below:
output:
<output qualifier> <output name>, emit: <output channel>This is very similar to the input block, except it can also have an emit: option.
The path qualifier specifies one or more files produced by the process into the specified channel as an output.
If you use single quotes (') around the output name, the name of the file outputted by the program has to match exactly to the one in the file path, otherwise it won't be collected in the process output.
Example
process RANDOMNUM {
output:
path 'result.txt'
script:
"""
echo \$RANDOM > result.txt
"""
}
workflow {
receiver_ch = RANDOMNUM()
receiver_ch.view()
}When an output file name contains a wildcard character (* or ?) it is interpreted as a glob path matcher. This allows us to capture multiple files into a list object and output them as a sole emission.
For example:
process SPLITLETTERS {
output:
path 'chunk_*'
script:
"""
printf 'Hola' | split -b 1 - chunk_
"""
}
workflow {
letters = SPLITLETTERS()
letters.view()
}Ouputs:
[/workspace/gitpod/nf-training/work/ca/baf931d379aa7fa37c570617cb06d1/chunk_aa, /workspace/gitpod/nf-training/work/ca/baf931d379aa7fa37c570617cb06d1/chunk_ab, /workspace/gitpod/nf-training/work/ca/baf931d379aa7fa37c570617cb06d1/chunk_ac, /workspace/gitpod/nf-training/work/ca/baf931d379aa7fa37c570617cb06d1/chunk_ad]
Some caveats on glob pattern behavior:
- Input files are not included in the list of possible matches
- Glob pattern matches both files and directory paths
- When a two stars pattern
**is used to recourse across directories, only file paths are matched i.e., directories are not included in the result list.
When an output file name needs to be expressed dynamically, it is possible to define it using a dynamic string that references values defined in the input declaration block or in the script global context.
For example:
species = ['cat', 'dog', 'sloth']
sequences = ['AGATAG', 'ATGCTCT', 'ATCCCAA']
Channel
.fromList(species)
.set { species_ch }
process ALIGN {
input:
val x
val seq
output:
path "${x}.aln"
script:
"""
echo align -in $seq > ${x}.aln
"""
}
workflow {
genomes = ALIGN(species_ch, sequences)
genomes.view()
}In the above example, each time the process is executed an alignment file is produced whose name depends on the actual value of the x input.
The ${..} syntax allows you to pass in input variable names to the other parts of the process.
A lot of examples show have used multiple input and output channels that can handle one value at a time. However, Nextflow can also handle a tuple of values.
The input and output declarations for tuples must be declared with a tuple qualifier followed by the definition of each element in the tuple.
This is really useful when you want to carry the ID of a sample through all the steps of your pipeline. The sample ID is something you'd want to carry through every step of your process so you can track what these files are. The files can change names after a lot of input and output steps but keeping them in a tuple with their sample ID will make it easier to figure out what they are. You might have different pieces of metadata being kept in the tuple as well.
Example:
reads_ch = Channel.fromFilePairs('data/ggal/*_{1,2}.fq')
process FOO {
input:
tuple val(sample_id), path(sample_id_paths)
output:
tuple val(sample_id), path('sample.bam')
script:
"""
echo your_command_here --sample $sample_id_paths > sample.bam
"""
}
workflow {
sample_ch = FOO(reads_ch)
sample_ch.view()
}Outputs:
[lung, /workspace/gitpod/nf-training/work/23/fe268295bab990a40b95b7091530b6/sample.bam]
[liver, /workspace/gitpod/nf-training/work/32/656b96a01a460f27fa207e85995ead/sample.bam]
[gut, /workspace/gitpod/nf-training/work/ae/3cfc7cf0748a598c5e2da750b6bac6/sample.bam]
Nextflow allows the use of alternative output definitions within workflows to simplify your code.
These are done with the .out attribute and the emit: option.
emit:is more commonly used than.outindexing
By using .out, your are getting the output channel of one process, and you can pass it in as the input channel of another process
Example
workflow {
reads_ch = Channel.fromFilePairs(params.reads)
prepare_star_genome_index(params.genome)
rnaseq_mapping_star(params.genome,
prepare_star_genome_index.out,
reads_ch)
}When a process defines multiple output channels, each output can be accessed using the array element operator (out[0], out[1], etc.)
Outputs can also be accessed by name if the emit option is specified
process example_process {
output:
path '*.bam', emit: samples_bam
'''
your_command --here
'''
}
workflow {
example_process()
example_process.out.samples_bam.view()
}If you have a process that only has a static pathname, for example
process rnaseq_call_variants {
container 'quay.io/broadinstitute/gotc-prod-gatk:1.0.0-4.1.8.0-1626439571'
tag "${sampleId}"
input:
path genome
path index
path dict
tuple val(sampleId), path(bam), path(bai)
output:
tuple val(sampleId), path('final.vcf')
script:
"""
...
"""This won't create a name conflict for every sample that gets processed.
This is because Nextflow creates a new directory for every task a process performs. So if you're trying to process 10 samples (run 10 tasks from a single process), you're going to have 10 isolated folders.
If you only had the path variable defined and not the tuple with the sampleId then it may have caused an issue but the way it's defined here file conflicts won't be an issue because every sample will get it's own folder.
Note
The Nextflow team suggests using a tuple with the ID attached to the sample instead of using the fair directive. You may experience some performance hits and less parallelism using the fair directive.
While channels do emit items in the order that they are received (FIFO structure), processes do not necessarily process items in the order that they are received (because of implicit parallelization and later processes ending before earlier ones). While this isn't an issue in most cases, it is important to know.
For example
process basicExample {
input:
val x
"echo process job $x"
}
workflow {
def num = Channel.of(1,2,3)
basicExample(num)
}Can output
process job 3
process job 1
process job 2
Notice in the above example that the value 3 was processed before the others.
The fair directive (new in version 22.12.0-edge), when enabled, guarantees that process outputs will be emitted in the order in which they were received. This is because the fair process directive distributes computing resources in a "fair" way (comes from fair-threading) to ensure the first one finishes first and so on.
Note
The Nextflow team suggests using a tuple with the ID attached to the sample instead of using the fair directive. You may experience some performance hits and less parallelism using the fair directive.
Example:
process EXAMPLE {
fair true
input:
val x
output:
tuple val(task.index), val(x)
script:
"""
sleep \$((RANDOM % 3))
"""
}
workflow {
channel.of('A','B','C','D') | EXAMPLE | view
}The above example produces:
[1, A]
[2, B]
[3, C]
[4, D]
The script block within a process is a string statement that defines the command to be executed by the process' task.
process CONVERTTOUPPER {
input:
path y
output:
stdout
script:
"""
cat $y | tr '[a-z]' '[A-Z]'
"""
}By default, Nextflow expects a shell script in the script block.
Note
Since Nextflow uses the same Bash syntax for variable substitutions in strings, Bash environment variables need to be escaped using the \ character. The escaped version will be resolved later, returning the task directory (e.g. work/7f/f285b80022d9f61e82cd7f90436aa4/), while $PWD would show the directory where you're running Nextflow.
Example:
process FOO {
debug true
script:
"""
echo "The current directory is \$PWD"
"""
}
workflow {
FOO()
}
// Outputs The current directory is /workspace/gitpod/nf-training/work/7a/4b050a6cdef4b6c1333ce29f7059a0And without \
process FOO {
debug true
script:
"""
echo "The current directory is $PWD"
"""
}
workflow {
FOO()
}
// The current directory is /workspace/gitpod/nf-trainingIt can be tricky to write a script that uses many Bash variables. One possible alternative is to use a script string delimited by single-quote characters (').
process BAR {
debug true
script:
'''
echo "The current directory is $PWD"
'''
}
workflow {
BAR()
}
// The current directory is /workspace/gitpod/nf-training/work/7a/4b050a6cdef4b6c1333ce29f7059a0However, this using the single quotes (') will block the usage of Nextflow variables in the command script.
Another alternative is to use a shell statement instead of script and use a different syntax for Nextflow variables, e.g., !{..}. This allows the use of both Nextflow and Bash variables in the same script.
Example:
params.data = 'le monde'
process BAZ {
shell:
'''
X='Bonjour'
echo $X !{params.data}
'''
}
workflow {
BAZ()
}If you are using another language, like R or Python, you need the shebang so that Nextflow knows which software to use to interpret this code.
Example:
process CONVERTTOUPPER {
input:
path y
output:
stdout
script:
"""
#!/usr/bin/env python
with open("$y") as f:
print(f.read().upper(), end="")
"""
}The process script can also be defined in a completely dynamic manner using an if statement or any other expression for evaluating a string value.
Example:
params.compress = 'gzip'
params.file2compress = "$baseDir/data/ggal/transcriptome.fa"
process FOO {
debug true
input:
path file
script:
if (params.compress == 'gzip')
"""
echo "gzip -c $file > ${file}.gz"
"""
else if (params.compress == 'bzip2')
"""
echo "bzip2 -c $file > ${file}.bz2"
"""
else
throw new IllegalArgumentException("Unknown compressor $params.compress")
}
workflow {
FOO(params.file2compress)
}Real-world workflows use a lot of custom user scripts (BASH, R, Python, etc.). Nextflow allows you to consistently use and manage these scripts. Simply put them in a directory named bin in the workflow project root. They will be automatically added to the workflow execution PATH.
For example, imagine this is a process block inside of main.nf
process FASTQ {
tag "FASTQ on $sample_id"
input:
tuple val(sample_id), path(reads)
output:
path "fastqc_${sample_id}_logs"
script:
"""
mkdir fastqc_${sample_id}_logs
fastqc -o fastqc_${sample_id}_logs -f fastq -q {reads}
"""
}The FASTQC process in main.nf could be replaced by creating an executable script named fastqc.sh in the bin directory as shown below:
Create a new file named fastqc.sh with the following content:
fastqc.sh
#!/bin/bash
set -e
set -u
sample_id=${1}
reads=${2}
mkdir fastqc_${sample_id}_logs
fastqc -o fastqc_${sample_id}_logs -f fastq -q ${reads}Give it execute permission and move it into the bin directory:
chmod +x fastqc.sh
mkdir -p bin
mv fastqc.sh binOpen the main.nf file and replace the FASTQC process script block with the following code:
main.nf
script:
"""
fastqc.sh "$sample_id" "$reads"
"""Channel factories are methods that can be used to create channels explicitly.
For example, the of method allows you to create a channel that emits the arguments provided to it, for example:
ch = Channel.of( 1, 3, 5, 7 )
ch.view { "value: $it" }The first line in this example creates a variable ch which holds a channel object. This channel emits the arguments supplied to the of method. Thus the second line prints the following:
value: 1
value: 3
value: 5
value: 7Channel factories also have options that can be used to modify their behaviour. For example, the checkIfExists option can be used to check if the specified path contains file pairs. If the path does not contain file pairs, an error is thrown. A full list of options can be found in the channel factory documentation.
The fromFilePairs method creates a channel emitting the file pairs matching a glob pattern provided by the user. The matching files are emitted as tuples in which the first element is the grouping key of the matching pair and the second element is the list of files (sorted in lexicographical order). For example:
Channel
.fromFilePairs('/my/data/SRR*_{1,2}.fastq')
.view()It will produce an output similar to the following:
[SRR493366, [/my/data/SRR493366_1.fastq, /my/data/SRR493366_2.fastq]]
[SRR493367, [/my/data/SRR493367_1.fastq, /my/data/SRR493367_2.fastq]]
[SRR493368, [/my/data/SRR493368_1.fastq, /my/data/SRR493368_2.fastq]]
[SRR493369, [/my/data/SRR493369_1.fastq, /my/data/SRR493369_2.fastq]]
[SRR493370, [/my/data/SRR493370_1.fastq, /my/data/SRR493370_2.fastq]]
[SRR493371, [/my/data/SRR493371_1.fastq, /my/data/SRR493371_2.fastq]]
The Channel.fromSRA channel factory makes it possible to query the NCBI SRA archive and returns a channel emitting the FASTQ files matching the specified selection criteria.
To learn more about how to use the fromSRA channel factory, see here.
Nextflow operators are methods that allow you to manipulate channels. Every operator, with the exception of set and subscribe, produces one or more new channels, allowing you to chain operators to fit your needs.
There are seven main groups of operators are described in greater detail within the Nextflow Reference Documentation, linked below:
- Filtering operators
- Transforming operators
- Splitting operators
- Combining operators
- Forking operators
- Maths operators
- Other operators
Sometimes the output channel of one process doesn't quite match the input channel of the next process and so it has to be modified slightly. This can be performed using channel operators. A full list of channel operators can be found here https://www.nextflow.io/docs/latest/operator.html.
For example, in this code:
rnaseq_gatk_analysis
.out
.groupTuple()
.join(prepare_vcf_for_ase.out.vcf_for_ASE)
.map { meta, bams, bais, vcf -> [meta, vcf, bams, bais] }
.set { grouped_vcf_bam_bai_ch }.groupTuplegroups tuples that contain a common first element.joinjoins two channels taking the key into consideration.mapapplies a function to every element of a channel.setsaves this channel with a new name
Step by step this looks like:
rnaseq_gatk_analysis
.out
/* Outputs
[ENCSR000COQ, /workspace/gitpod/hands-on/work/c9/dfd66e253754b61195a166ac7726ff/ENCSR000COQ1.final.uniq.bam, /workspace/gitpod/hands-on/work/c9/dfd66e253754b61195a166ac7726ff/ENCSR000COQ1.final.uniq.bam.bai]
[ENCSR000COQ, /workspace/gitpod/hands-on/work/c9/dfd66e253754b61195a166ac7726ff/ENCSR000COQ2.final.uniq.bam, /workspace/gitpod/hands-on/work/c9/dfd66e253754b61195a166ac7726ff/ENCSR000COQ2.final.uniq.bam.bai]
*/
.groupTuple()
/* Outputs
[ENCSR000COQ, [/workspace/gitpod/hands-on/work/c9/dfd66e253754b61195a166ac7726ff/ENCSR000COQ1.final.uniq.bam, /workspace/gitpod/hands-on/work/92/b1ea340ce922d13bdce2985b2930f2/ENCSR000COQ2.final.uniq.bam], [/workspace/gitpod/hands-on/work/c9/dfd66e253754b61195a166ac7726ff/ENCSR000COQ1.final.uniq.bam.bai, /workspace/gitpod/hands-on/work/92/b1ea340ce922d13bdce2985b2930f2/ENCSR000COQ2.final.uniq.bam.bai]]
*/
.join(prepare_vcf_for_ase.out.vcf_for_ASE)
/* Outputs
[ENCSR000COQ, [/workspace/gitpod/hands-on/work/c9/dfd66e253754b61195a166ac7726ff/ENCSR000COQ1.final.uniq.bam, /workspace/gitpod/hands-on/work/92/b1ea340ce922d13bdce2985b2930f2/ENCSR000COQ2.final.uniq.bam], [/workspace/gitpod/hands-on/work/c9/dfd66e253754b61195a166ac7726ff/ENCSR000COQ1.final.uniq.bam.bai, /workspace/gitpod/hands-on/work/92/b1ea340ce922d13bdce2985b2930f2/ENCSR000COQ2.final.uniq.bam.bai], /workspace/gitpod/hands-on/work/ea/4a41fbeb591ffe48cfb471890b8f5c/known_snps.vcf]
*/
.map { meta, bams, bais, vcf -> [meta, vcf, bams, bais] }
/* Outputs
[ENCSR000COQ, /workspace/gitpod/hands-on/work/ea/4a41fbeb591ffe48cfb471890b8f5c/known_snps.vcf, [/workspace/gitpod/hands-on/work/c9/dfd66e253754b61195a166ac7726ff/ENCSR000COQ1.final.uniq.bam, /workspace/gitpod/hands-on/work/92/b1ea340ce922d13bdce2985b2930f2/ENCSR000COQ2.final.uniq.bam], [/workspace/gitpod/hands-on/work/c9/dfd66e253754b61195a166ac7726ff/ENCSR000COQ1.final.uniq.bam.bai, /workspace/gitpod/hands-on/work/92/b1ea340ce922d13bdce2985b2930f2/ENCSR000COQ2.final.uniq.bam.bai]]
*/
.set { grouped_vcf_bam_bai_ch }Just keep in mind processes and channel operators are not guaranteed to emit items in the order that they were received, as they are executed concurrently. This can lead to unintended effects based if you use a operator that takes multiple inputs. For example, the using the merge channel operator can lead to different results upon different runs based on the order in which the processes finish. You should always use a matching key (e.g. sample ID) to merge multiple channels, so that they are combined in a deterministic way (using an operator like join instead).
The view operator prints the items emitted by a channel to the console standard output, appending a new line character to each item.
For example:
Channel
.of('foo', 'bar', 'baz')
.view()Outputs:
foo
bar
baz
An optional closure parameter can be specified to customize how items are printed.
For example:
Channel
.of('foo', 'bar', 'baz')
.view { "- $it" }- foo
- bar
- baz
Note
The closure replaced the () brackets in the view operator with {} brackets.
The map operator applies a function of your choosing to every item emitted by a channel and returns the items obtained as a new channel. The function applied is called the mapping function and is expressed with a closure.
In the example below the groovy reverse method has been used to reverse the order of the characters in each string emitted by the channel.
Channel
.of('hello', 'world')
.map { it -> it.reverse() }
.view()Outputs:
olleh
dlrow
A map can associate a generic tuple to each element and can contain any data.
In the example below the groovy size method is used to return the length of each string emitted by the channel.
Channel
.of('hello', 'world')
.map { word -> [word, word.size()] }
.view()Outputs:
[hello, 5]
[world, 5]
The first operator emits the first item in a source channel, or the first item that matches a condition, and outputs a value channel. The condition can be a regular expression, a type qualifier (i.e. Java class), or a boolean predicate.
For example:
// no condition is specified, emits the very first item: 1
Channel.of( 1, 2, 3 )
.first()
.view()
// emits the first item matching the regular expression: 'aa'
Channel.of( 'a', 'aa', 'aaa' )
.first( ~/aa.*/ )
.view()
// emits the first String value: 'a'
Channel.of( 1, 2, 'a', 'b', 3 )
.first( String )
.view()
// emits the first item for which the predicate evaluates to true: 4
Channel.of( 1, 2, 3, 4, 5 )
.first { it > 3 }
.view()Since this returns a value channel and value channels are inexaustable, this is really good for channels that contain files which are reused oftens, like reference genomes.
Example:
reference = Channel.fromPath("data/genome.fasta").first()The .flatten operator transforms a channel in such a way that every item of type Collection or Array is flattened so that each single entry is emitted separately by the resulting channel. For example:
Channel
.of( [1,[2,3]], 4, [5,[6]] )
.flatten()
.view()Outputs
1
2
3
4
5
6
Note
This is different to .mix because .mix operates on items emitted from channels, not Collection or Array objects.
The .collect channel operator is basically the oposite of the .flatten channel operator, where it collects all the items emitted by a channel to a List and return the resulting object as a sole emission. For example:
Channel
.of( 1, 2, 3, 4 )
.collect()
.view()Outputs
[1,2,3,4]
By default, .collect will flatten nested list objects and collect their items individually. To change this behaviour, set the flat option to false.
Example:
Channel
.of( [1, 2], [3, 4] )
.collect()
.view()
// Outputs
// [1,2,3,4]
Channel
.of( [1, 2], [3, 4] )
.collect(flat: false)
.view()
// Outputs
// [[1,2],[3,4]]The .collect operator also can take a sort option (false by default). sort
When true, the collected items are sorted by their natural ordering. Can also be a closure or a Comparator which defines how items are compared during sorting.
The .buffer channel operator gathers the items emitted by the source channel into subsets and emits these subsets separately.
There are a number of ways you can regulate how buffer gathers the items from the source channel into subsets, however one of the most convienient ways of using it is with buffer( size: n ). transform the source channel in such a way that it emits tuples made up of n elements. For example:
Channel
.of( 1, 2, 3, 1, 2, 3, 1 )
.buffer( size: 2 )
.view()Outputs
[1, 2]
[3, 1]
[2, 3]
Be aware that if there is an incomplete tuple it is discarded. To emit the last items in a tuple containing less than n elements, use buffer( size: n, remainder: true ). For example:
Channel
.of( 1, 2, 3, 1, 2, 3, 1 )
.buffer( size: 2, remainder: true )
.view()Outputs
[1, 2]
[3, 1]
[2, 3]
[1]
The mix operator combines the items emitted by two (or more) channels into a single queue channel.
For example:
c1 = Channel.of( 1, 2, 3 )
c2 = Channel.of( 'a', 'b' )
c3 = Channel.of( 'z' )
c1.mix(c2,c3)
.subscribe onNext: { println it }, onComplete: { println 'Done' }Outputs:
1
2
3
'a'
'b'
'z'
Note
The items emitted by the resulting mixed channel may appear in any order, regardless of which source channel they came from. Thus, the following example could also be a possible result of the above example:
'z'
1
'a'
2
'b'
3
The mix operator and the collect operator are often chained together to gather the outputs of the multiple processes as a single input. Operators can be used in combinations to combine, split, and transform channels.
Example:
MULTIQC(quant_ch.mix(fastqc_ch).collect())You will only want one task of MultiQC to be executed to produce one report. Therefore, you can use the mix channel operator to combine the quant_ch and the fastqc_ch channels, followed by the collect operator, to return the complete channel contents as a single element.
Note
mix is different to .flatten because flatten operates on Collection or Array objects, not individual items.
Things can get quite complicated when you have lots of different tuples, especially when you're trying to use something like matching keys.
For example, if you had a sample with a bunch of different chromosomes, and you wanted to split them up all together and process them individually. How would you merge them back together based on a sample ID?
The groupTuple operator is useful for this.
The groupTuple operator collects tuples (or lists) of values emitted by the source channel, grouping the elements that share the same key into a list afterwards. Finally, it emits a new tuple object for each distinct key collected.
By default, the first element of each tuple is used as the grouping key. The by option can be used to specify a different index, or list of indices. See here for more details.
Channel
.of([1, 'A'], [1, 'B'], [2, 'C'], [3, 'B'], [1, 'C'], [2, 'A'], [3, 'D'])
.groupTuple()
.view()Outputs
[1, [A, B, C]]
[2, [C, A]]
[3, [B, D]]
groupTuple is very useful alongside 'meta maps' (see here for example)
Note
By default, groupTuple is a blocking operator, meanining it won't emit anything until all the inputs have been adjusted (see example above for further explaination).
By default, if you don’t specify a size, the groupTuple operator will not emit any groups until all inputs have been received. If possible, you should always try to specify the number of expected elements in each group using the size option, so that each group can be emitted as soon as it’s ready. In cases where the size of each group varies based on the grouping key, you can use the built-in groupKey() function, which allows you to define a different expected size for each group:
chr_frequency = ["chr1": 2, "chr2": 3]
Channel.of(
['region1', 'chr1', '/path/to/region1_chr1.vcf'],
['region2', 'chr1', '/path/to/region2_chr1.vcf'],
['region1', 'chr2', '/path/to/region1_chr2.vcf'],
['region2', 'chr2', '/path/to/region2_chr2.vcf'],
['region3', 'chr2', '/path/to/region3_chr2.vcf']
)
.map { region, chr, vcf -> tuple( groupKey(chr, chr_frequency[chr]), vcf ) }
.groupTuple()
.view()Outputs:
[chr1, [/path/to/region1_chr1.vcf, /path/to/region2_chr1.vcf]]
[chr2, [/path/to/region1_chr2.vcf, /path/to/region2_chr2.vcf, /path/to/region3_chr2.vcf]]
As a further explaination, take a look at the following example which shows a dummy read mapping process.:
process MapReads {
input:
tuple val(meta), path(reads)
path(genome)
output:
tuple val(meta), path("*.bam")
"touch out.bam"
}
workflow {
reference = Channel.fromPath("data/genome.fasta").first()
Channel.fromPath("data/samplesheet.csv")
| splitCsv( header:true )
| map { row ->
meta = row.subMap('id', 'repeat', 'type')
[meta, [
file(row.fastq1, checkIfExists: true),
file(row.fastq2, checkIfExists: true)]]
}
| set { samples }
MapReads( samples, reference )
| view
}Let's consider that you might now want to merge the repeats. You'll need to group bams that share the id and type attributes.
MapReads( samples, reference )
| map { meta, bam -> [meta.subMap('id', 'type'), bam]}
| groupTuple
| viewThis is easy enough, but the groupTuple operator has to wait until all items are emitted from the incoming queue before it is able to reassemble the output queue. If even one read mapping job takes a long time, the processing of all other samples is held up. You need a way of signalling to Nextflow how many items are in a given group so that items can be emitted as early as possible.
By default, the groupTuple operator groups on the first item in the element, which at the moment is a Map. You can turn this map into a special class using the groupKey method, which takes our grouping object as a first parameter and the number of expected elements in the second parameter.
MapReads( samples, reference )
| map { meta, bam ->
key = groupKey(meta.subMap('id', 'type'), NUMBER_OF_ITEMS_IN_GROUP)
[key, bam]
}
| groupTuple
| viewSo modifying the upstream channel would look like this:
workflow {
reference = Channel.fromPath("data/genome.fasta").first()
Channel.fromPath("data/samplesheet.csv")
| splitCsv( header:true )
| map { row ->
meta = row.subMap('id', 'repeat', 'type')
[meta, [file(row.fastq1, checkIfExists: true), file(row.fastq2, checkIfExists: true)]]
}
| map { meta, reads -> [meta.subMap('id', 'type'), meta.repeat, reads] }
| groupTuple
| map { meta, repeats, reads -> [meta + [repeatcount:repeats.size()], repeats, reads] }
| transpose
| map { meta, repeat, reads -> [meta + [repeat:repeat], reads]}
| set { samples }
MapReads( samples, reference )
| map { meta, bam ->
key = groupKey(meta.subMap('id', 'type'), meta.repeatcount)
[key, bam]
}
| groupTuple
| view
}Now that you have our repeats together in an output channel, you can combine them using "advanced bioinformatics":
process CombineBams {
input:
tuple val(meta), path("input/in_*_.bam")
output:
tuple val(meta), path("combined.bam")
"cat input/*.bam > combined.bam"
}
// ...
// In the workflow
MapReads( samples, reference )
| map { meta, bam ->
key = groupKey(meta.subMap('id', 'type'), meta.repeatcount)
[key, bam]
}
| groupTuple
| CombineBamsSo because here groupKey is being used, the elements from groupTuple were emitted much faster than they would otherwise have been because you don't have to wait for all of the mapping operations to complete before your groupTuple operation starts to emit items. The groupTuple operator already knows that some of these samples are ready because as soon as the second argument of groupKey is satisfied (in this case the length of the meta.repeatcount value) it knows that the tuple is ready to be emitted and will emit it immediately instead of having to wait for all the samples. This is very useful for when you have large runs with tens to hundreds of samples and will save lots of time by emitting ready items as soon as possible for downstream process to begin working on them.
Note
The class of a groupKey object is nextflow.extension.GroupKey. If you need the raw contents of the groupKey object (e.g. the metadata map), you can use the .getGroupTarget() method to extract those. See here for more information.
The transpose operator is often misunderstood. It can be thought of as the inverse of the groupTuple operator (see here for an example).
Given the following workflow, the groupTuple and transpose operators cancel each other out. Removing lines 8 and 9 returns the same result.
Given a workflow that returns one element per sample, where we have grouped the samplesheet lines on a meta containing only id and type:
workflow {
Channel.fromPath("data/samplesheet.csv")
| splitCsv(header: true)
| map { row ->
meta = [id: row.id, type: row.type]
[meta, row.repeat, [row.fastq1, row.fastq2]]
}
| groupTuple
| view
}Outputs:
N E X T F L O W ~ version 23.04.1
Launching `./main.nf` [spontaneous_rutherford] DSL2 - revision: 7dc1cc0039
[[id:sampleA, type:normal], [1, 2], [[data/reads/sampleA_rep1_normal_R1.fastq.gz, data/reads/sampleA_rep1_normal_R2.fastq.gz], [data/reads/sampleA_rep2_normal_R1.fastq.gz, data/reads/sampleA_rep2_normal_R2.fastq.gz]]]
[[id:sampleA, type:tumor], [1, 2], [[data/reads/sampleA_rep1_tumor_R1.fastq.gz, data/reads/sampleA_rep1_tumor_R2.fastq.gz], [data/reads/sampleA_rep2_tumor_R1.fastq.gz, data/reads/sampleA_rep2_tumor_R2.fastq.gz]]]
[[id:sampleB, type:normal], [1], [[data/reads/sampleB_rep1_normal_R1.fastq.gz, data/reads/sampleB_rep1_normal_R2.fastq.gz]]]
[[id:sampleB, type:tumor], [1], [[data/reads/sampleB_rep1_tumor_R1.fastq.gz, data/reads/sampleB_rep1_tumor_R2.fastq.gz]]]
[[id:sampleC, type:normal], [1], [[data/reads/sampleC_rep1_normal_R1.fastq.gz, data/reads/sampleC_rep1_normal_R2.fastq.gz]]]
[[id:sampleC, type:tumor], [1], [[data/reads/sampleC_rep1_tumor_R1.fastq.gz, data/reads/sampleC_rep1_tumor_R2.fastq.gz]]]
If we add in a transpose, each repeat number is matched back to the appropriate list of reads:
workflow {
Channel.fromPath("data/samplesheet.csv")
| splitCsv(header: true)
| map { row ->
meta = [id: row.id, type: row.type]
[meta, row.repeat, [row.fastq1, row.fastq2]]
}
| groupTuple
| transpose
| view
}Outputs:
N E X T F L O W ~ version 23.04.1
Launching `./main.nf` [elegant_rutherford] DSL2 - revision: 2c5476b133
[[id:sampleA, type:normal], 1, [data/reads/sampleA_rep1_normal_R1.fastq.gz, data/reads/sampleA_rep1_normal_R2.fastq.gz]]
[[id:sampleA, type:normal], 2, [data/reads/sampleA_rep2_normal_R1.fastq.gz, data/reads/sampleA_rep2_normal_R2.fastq.gz]]
[[id:sampleA, type:tumor], 1, [data/reads/sampleA_rep1_tumor_R1.fastq.gz, data/reads/sampleA_rep1_tumor_R2.fastq.gz]]
[[id:sampleA, type:tumor], 2, [data/reads/sampleA_rep2_tumor_R1.fastq.gz, data/reads/sampleA_rep2_tumor_R2.fastq.gz]]
[[id:sampleB, type:normal], 1, [data/reads/sampleB_rep1_normal_R1.fastq.gz, data/reads/sampleB_rep1_normal_R2.fastq.gz]]
[[id:sampleB, type:tumor], 1, [data/reads/sampleB_rep1_tumor_R1.fastq.gz, data/reads/sampleB_rep1_tumor_R2.fastq.gz]]
[[id:sampleC, type:normal], 1, [data/reads/sampleC_rep1_normal_R1.fastq.gz, data/reads/sampleC_rep1_normal_R2.fastq.gz]]
[[id:sampleC, type:tumor], 1, [data/reads/sampleC_rep1_tumor_R1.fastq.gz, data/reads/sampleC_rep1_tumor_R2.fastq.gz]]
The join operator is very similar to the groupTuple operator except it joins elements from multiple channels.
The join operator creates a channel that joins together the items emitted by two channels with a matching key. The key is defined, by default, as the first element in each item emitted.
left = Channel.of(['X', 1], ['Y', 2], ['Z', 3], ['P', 7])
right = Channel.of(['Z', 6], ['Y', 5], ['X', 4])
left.join(right).view()Output
[Z, 3, 6]
[Y, 2, 5]
[X, 1, 4]
Note
by default, join drops elements that don't have a match (Notice the P key and its corresponding list elements in the above example is missing from the output). This behaviour can be changed with the remainder option. See here for more details.
The branch operator allows you to forward the items emitted by a source channel to one or more output channels.
The selection criterion is defined by specifying a closure that provides one or more boolean expressions, each of which is identified by a unique label. For the first expression that evaluates to a true value, the item is bound to a named channel as the label identifier.
Example:
Channel
.of(1, 2, 3, 10, 40, 50)
.branch {
small: it <= 10
large: it > 10
}
.set { result }
result.small.view { "$it is small" }
result.large.view { "$it is large" }Output
1 is small
40 is large
2 is small
10 is small
3 is small
50 is large
An element is only emitted to a channel where the test condition is met. If an element does not meet any of the tests, it is not emitted to any of the output channels. You can 'catch' any such samples by specifying true as a condition.
Example:
workflow {
Channel.fromPath("data/samplesheet.csv")
| splitCsv( header: true )
| map { row -> [[id: row.id, repeat: row.repeat, type: row.type], [file(row.fastq1), file(row.fastq2)]] }
| branch { meta, reads ->
tumor: meta.type == "tumor"
normal: meta.type == "normal"
other: true
}
| set { samples }
samples.tumor | view { "Tumor: $it"}
samples.normal | view { "Normal: $it"}
samples.other | view { log.warn "Non-tumour or normal sample found in samples: $it"}
}other here will catch any rows that don't fall into either branches. This is good for testing for typos or errors in the data.
You can then combine this with log functions log.warn to warn the user there may be an error or more strictly log.error to halt the pipeline.
You may also want to emit a slightly different element than the one passed as input. The branch operator can (optionally) return a new element to an channel. For example, to add an extra key in the meta map of the tumor samples, we add a new line under the condition and return our new element. In this example, we modify the first element of the List to be a new list that is the result of merging the existing meta map with a new map containing a single key:
branch { meta, reads ->
tumor: meta.type == "tumor"
return [meta + [newKey: 'myValue'], reads]
normal: true
}The multiMap channel operator is similar to the branch channel operator
The multiMap (documentation) operator is a way of taking a single input channel and emitting into multiple channels for each input element.
Let's assume we've been given a samplesheet that has tumor/normal pairs bundled together on the same row. View the example samplesheet with:
cd operators
cat data/samplesheet.ugly.csvUsing the splitCsv operator would give us one entry that would contain all four fastq files. Let's consider that we wanted to split these fastqs into separate channels for tumor and normal. In other words, for every row in the samplesheet, we would like to emit an entry into two new channels. To do this, we can use the multiMap operator:
workflow {
Channel.fromPath("data/samplesheet.ugly.csv")
| splitCsv( header: true )
| multiMap { row ->
tumor:
metamap = [id: row.id, type:'tumor', repeat:row.repeat]
[metamap, file(row.tumor_fastq_1), file(row.tumor_fastq_2)]
normal:
metamap = [id: row.id, type:'normal', repeat:row.repeat]
[metamap, file(row.normal_fastq_1), file(row.normal_fastq_2)]
}
| set { samples }
samples.tumor | view { "Tumor: $it"}
samples.normal | view { "Normal: $it"}
}One way to think about this is if you have for example data in a .csv file.
- If you need to output two channels for every single row, use
multiMap - If you need to output two channels, and you need to filter which parts of all the rows should go into which channel, use
branch
The flatMap operator allows you to modify the elements in a channel and then flatten the resulting collection. This is useful if you need to "expand" elements in a channel an incoming element can turn into zero or more elements in the output channel.
For example:
workflow {
numbers = Channel.of(1, 2)
numbers
| flatMap { n -> [ n, n*10, n*100 ] }
| view
}The input channel has two elements. For each element in the input channel, we return a List of length three. The List is flattened and each element in our returned list is emitted independently into the output channel:
Outputs:
1
10
100
2
20
200
This differs from the flatten operator because the flatten operator only "unfolds" one layer from the returned collection
The combine operator produces the combinations (i.e. cross product, “Cartesian” product) of two source channels, or a channel and a list (as the right operand), emitting each combination separately.
For example:
numbers = Channel.of(1, 2, 3)
words = Channel.of('hello', 'ciao')
numbers
.combine(words)
.view()Outputs:
[1, hello]
[2, hello]
[3, hello]
[1, ciao]
[2, ciao]
[3, ciao]
The by option can be used to combine items that share a matching key. The value should be the zero-based index of the tuple, or a list of indices.
For example:
source = Channel.of( [1, 'alpha'], [2, 'beta'] )
target = Channel.of( [1, 'x'], [1, 'y'], [1, 'z'], [2, 'p'], [2, 'q'], [2, 't'] )
source.combine(target, by: 0).view()Outputs:
[1, alpha, x]
[1, alpha, y]
[1, alpha, z]
[2, beta, p]
[2, beta, q]
[2, beta, t]
This is very useful for splitting or fanning-out your data to perforom operations on multiple subgroups of your data (See here for more info).
Note
The combine operator is similar to cross and join, making them easy to confuse. Their differences can be summarized as follows:
combineandcrossboth produce an outer product or cross product, whereasjoinproduces an inner product.combinefilters pairs with a matching key only if the by option is used, whereascrossalways filters pairs with a matching key.combinewith the by option merges and flattens each pair, whereascrossdoes not. Compare the examples here forcombineandcrossto see this difference.
The set operator assigns the channel to a variable whose name is specified as a closure parameter. It is used in place of the assignment (=) operator. For example:
Channel.of(10, 20, 30).set { my_channel }This is semantically equivalent to the following assignment:
my_channel = Channel.of(10, 20, 30)However the set operator is more grammatical in Nextflow scripting, since it can be used at the end of a chain of operator transformations, thus resulting in a more fluent and readable operation.
Whichever way you choose to assign a variable is up to you.
The collectFile operator allows you to write one or more new files based on the contents of a channel.
More information on how this works can be found here.
Nextflow also offers a lot of channel operators for working with common text file formats (e.g. .txt, .csv, .json). More information on these and how they work can be found here.
Sometimes you'll have a filepath you want to use in your workflow (e.g. path/to/some/file). It's a good idea to check the class of your file because it may be a string instead of a file object.
Turning your file path into a path object is important because it helps Nextflow provision and stage your data.
Example:
SOME_PROCESS.out.view()
// outputs path/to/some/file
SOME_PROCESS.out.view{ it.getClass() }
// outputs java.lang.String
SOME_PROCESS.out.view{ file(it).getClass() }
// outputs sun.nio.fs.UnixPathThe file method also comes with an optional checkIfExists parameter, that will check if the file exists. It works on local machines and file systems on remote files and block storage.
Example:
workflow {
Channel.fromPath("data/samplesheet.csv")
| splitCsv( header:true )
| map { row ->
meta = row.subMap('id', 'repeat', 'type')
[meta, [
file(row.fastq1, checkIfExists: true),
file(row.fastq2, checkIfExists: true)]]
}
| view
}Ranges of values are expanded accordingly:
Channel
.of(1..23, 'X', 'Y')
.view()Prints:
1
2
3
4
:
23
X
YInputs and outputs in Nextflow need to a data type assigned before a variable name. If the data type is a tuple, all the items in the tuple need a data type as well.
Example
process example_process {
input:
tuple val(replicateId), path(reads)
output:
tuple val(replicateId),
path('Aligned.sortedByCoord.out.bam'),
path('Aligned.sortedByCoord.out.bam.bai')
script:
"""
...
"""
}Value channels allow processes to consume elements infinite times.
Elements in the queue channel are consumed. You cannot consume the same element for the same process twice.
One common use case of a value channel is when you're working with a reference genome. Often times you'll want to map many reads back to the reference genome but you don't want the reference genome to get consumed on the first mapping. Therefore be aware of what inputs you want to reuse over and over again and which inputs you want to consume.
For more information, see here.
Note
If two different processes require the same input, the channel is automatically duplicated if you provide it as input for multiple processes. This is a new feature of the DSL2 language update. Earlier you indeed had to manually duplicate the queue channel in such cases. The distinction between value and queue channels now is mostly relevant for processes with multiple input channels.
Processes can only take channels as inputs. That being said, if you pass in a regular variable that has a single value (e.g. string, number, etc), Nextflow will implicitly create a value channel containing that value that you are providing. Therefore you can pass a regular value (i.e. string, number) into a process, but just be aware that this is what is going on behind the scenes
Example
/*
* Define the default parameters
*/
params.genome = "${baseDir}/data/genome.fa"
params.variants = "${baseDir}/data/known_variants.vcf.gz"
params.blacklist = "${baseDir}/data/blacklist.bed"
params.reads = "${baseDir}/data/reads/ENCSR000COQ1_{1,2}.fastq.gz"
params.results = "results"
/*
* Process 1A: Create a FASTA genome index with samtools
*/
process prepare_genome_samtools {
container 'quay.io/biocontainers/samtools:1.3.1--h0cf4675_11'
input:
path genome
output:
path "${genome}.fai"
script:
"""
samtools faidx ${genome}
"""
}
workflow {
reads_ch = Channel.fromFilePairs(params.reads)
prepare_genome_samtools(params.genome)
}The prepare_genome_samtools(params.genome) is a valid call to a process because params.genome will be converted from a string into a value channel.
By default, the task work directories are created in the directory from where the pipeline is launched. This is often a scratch storage area that can be cleaned up once the computation is completed. A different location for the execution work directory can be specified using the command line option -w
Example
nextflow run <script> -w /some/scratch/dirNote
If you delete or move the pipeline work directory, this will prevent to use the resume feature in subsequent runs.
Note
The pipeline work directory is intended to be used as a temporary scratch area. The final workflow outputs are expected to be stored in a different location specified using the publishDir directive.
In order to use the processDir directive to publish your desired files to some output directory, add the following to your process block
process SOMEPROCESS {
publishDir params.outdir, mode: 'copy'
}You want to provide the mode: 'copy' option because by default files are published to the target folder creating a symbolic link for each process output that links the file produced into the process working directory. Usually you want an actual copy of your desired file and not just a symbolic link.
You can also provide the option saveAs to the publishDir directive if you want to rename the names of the output files. saveAs takes in a closure. If you want to not publish a file (e.g. if you only want to publish some output files from a process that produces multiple of them), return the value null from the closure.
Example (this example is using the syntax found in config files):
process {
withName: 'FASTP' {
publishDir = [
path: { "results/fastp/json" },
saveAs: { filename -> filename.endsWith('.json') ? filename : null },
]
}
}Note
As of Nextflow v24.04, the workflow output definition is the new preferred style for publishing output files.
The workflow output definition is a new syntax for defining workflow outputs. For example:
nextflow.preview.output = true
workflow {
main:
ch_foo = foo(data)
bar(ch_foo)
publish:
ch_foo >> 'foo'
}
output {
directory 'results'
mode 'copy'
} This essentially provides a DSL2-style approach for publishing, and will replace publishDir once it is finalized. It also provides extra flexibility as it allows you to publish any channel, not just process outputs. See the Nextflow docs for more information.
Warning
As of right now, this feature is still in preview and may change in a future release. The Seqera teams hopes to finalize it in version 24.10, so be aware of any issues that may come up.
You can set up an event handler in your workflow to do something when the script finishes using the workflow.onComplete event handler.
Example:
workflow.onComplete {
log.info ( worflow.success ? "\nDone! Open the following report in your browser --> $params.outdir/multiqc_report.html\n" : 'Oops .. something went wrong' )
}Nextflow DSL2 allows for the definition of stand-alone module scripts that can be included and shared across multiple workflows. Each module can contain its own process or workflow definition.
Commonly modules are stored in main.nf files within a module folder and are imported the top-level main.nf file.
Example:
.
├── main.nf
└── modules
└── local
└── fastp
└── main.nf
The module is then imported into the top-level main.nf using:
include { FASTP } from './modules/local/fastp/main.nf'More information can be found here (Take careful note of the Module aliases section. This section talks about how to invoke processes multiple times after importing).
nf-core provides some pre-built modules that can be useful (see here).
These can also be viewed from the command line if you have nf-core installed.
# If you don't have it installed
conda activate mamba
mamba install nf-core
# View the available modules
nf-core modules
# Get help installing a module
nf-core modules install --help
# Example: Install FASTQC to your pipeline directory
nfcore modules install . fastqcTo use a nf-core module (and a local one too), you should:
- Update the
conf/modules.configfile. - Add
includeConfig 'conf/modules.config'tonextflow.config. - Include module into workflows and/or subworkflows.
More information on nf-core modules can be found in this video tutorial.
Caution
Modules in the modules/nf-core directory can be synced with the modules in a remote repository. Any local changes or edits made to these can be lost if you're not careful. For more information see this video.
MultiQC is a tool used for the aggregation and summarization of bioinformatics analysis results.
MultiQC simplifies the process of managing and interpreting the results from various bioinformatics analysis tools by automatically collecting and summarizing the outputs from multiple tools into a single, easy-to-read HTML report. It identifies common metrics and trends across different analysis tools, providing a comprehensive overview of the data analysis process.
However as a module, MULTIQC is special because it has to be made local. You have to customize it for all the tools and modules you used for your workflows and subworkflows.
Each module should (and must if in the context of nf-core) emit the software versions of their tools.
The workflow scope allows the definition of components that define the invocation of one or more processes or operators.
Example:
#!/usr/bin/env nextflow
params.greeting = 'Hello world!'
include { SPLITLETTERS } from './modules.nf'
include { CONVERTTOUPPER } from './modules.nf'
workflow my_workflow {
greeting_ch = Channel.of(params.greeting)
SPLITLETTERS(greeting_ch)
CONVERTTOUPPER(SPLITLETTERS.out.flatten())
CONVERTTOUPPER.out.view { it }
}
workflow {
my_workflow()
}The snippet above defines a workflow named my_workflow, that is invoked via another workflow definition.
A workflow component can declare one or more input channels using the take statement.
For example:
#!/usr/bin/env nextflow
params.greeting = 'Hello world!'
include { SPLITLETTERS } from './modules.nf'
include { CONVERTTOUPPER } from './modules.nf'
workflow my_workflow {
take:
greeting
main:
SPLITLETTERS(greeting)
CONVERTTOUPPER(SPLITLETTERS.out.flatten())
CONVERTTOUPPER.out.view { it }
}The input for the workflow can then be specified as an argument:
workflow {
my_workflow(Channel.of(params.greeting))
}A workflow can declare one or more output channels using the emit statement.
For example:
#!/usr/bin/env nextflow
params.greeting = 'Hello world!'
greeting_ch = Channel.of(params.greeting)
process SPLITLETTERS {
input:
val x
output:
path 'chunk_*'
"""
printf '$x' | split -b 6 - chunk_
"""
}
process CONVERTTOUPPER {
input:
path y
output:
stdout emit: upper
"""
cat $y | tr '[a-z]' '[A-Z]'
"""
}
workflow my_workflow {
take:
greeting
main:
SPLITLETTERS(greeting)
CONVERTTOUPPER(SPLITLETTERS.out.flatten())
emit:
CONVERTTOUPPER.out.upper
}
workflow {
my_workflow(Channel.of(params.greeting))
my_workflow.out.view()
}As a result, you can use the my_workflow.out notation to access the outputs of my_workflow in the invoking workflow.
You can also declare named outputs within the emit block.
Example:
#!/usr/bin/env nextflow
params.greeting = 'Hello world!'
greeting_ch = Channel.of(params.greeting)
process SPLITLETTERS {
input:
val x
output:
path 'chunk_*'
"""
printf '$x' | split -b 6 - chunk_
"""
}
process CONVERTTOUPPER {
input:
path y
output:
stdout emit: upper
"""
cat $y | tr '[a-z]' '[A-Z]'
"""
}
workflow my_workflow {
take:
greeting
main:
SPLITLETTERS(greeting)
CONVERTTOUPPER(SPLITLETTERS.out.flatten())
emit:
my_data = CONVERTTOUPPER.out.upper
}
workflow {
my_workflow(Channel.of(params.greeting))
my_workflow.out.my_data.view()
}The result of the above snippet can then be accessed using my_workflow.out.my_data.
Within a main.nf script you can also have multiple workflows. In which case you may want to call a specific workflow when running the code. For this you could use the entrypoint call -entry <workflow_name>.
The following snippet has two named workflows (my_workflow_one and my_workflow_two):
#!/usr/bin/env nextflow
params.greeting = 'Hello world!'
include { SPLITLETTERS as SPLITLETTERS_one } from './modules.nf'
include { SPLITLETTERS as SPLITLETTERS_two } from './modules.nf'
include { CONVERTTOUPPER as CONVERTTOUPPER_one } from './modules.nf'
include { CONVERTTOUPPER as CONVERTTOUPPER_two } from './modules.nf'
workflow my_workflow_one {
letters_ch1 = SPLITLETTERS_one(params.greeting)
results_ch1 = CONVERTTOUPPER_one(letters_ch1.flatten())
results_ch1.view { it }
}
workflow my_workflow_two {
letters_ch2 = SPLITLETTERS_two(params.greeting)
results_ch2 = CONVERTTOUPPER_two(letters_ch2.flatten())
results_ch2.view { it }
}
workflow {
my_workflow_one(Channel.of(params.greeting))
my_workflow_two(Channel.of(params.greeting))
}You can choose which workflow to run by using the entry flag:
nextflow run hello2.nf -entry my_workflow_oneNextflow can produce multiple reports and charts providing several runtime metrics and execution information. These can be enabled by using the following command line options:
The -with-report option enables the creation of the workflow execution report.
The -with-trace option enables the creation of a tab separated value (TSV) file containing runtime information for each executed task.
The -with-timeline option enables the creation of the workflow timeline report showing how processes were executed over time. This may be useful to identify the most time consuming tasks and bottlenecks.
Finally, the -with-dag option enables the rendering of the workflow execution direct acyclic graph representation. The dag needs to be given a name (-with-dag dag.png).
Note
This feature requires the installation of Graphviz on your computer. See here for further details. You can also output HTML DAGs (-with-dag dag.html), which means Nextflow is going to use Mermaid for rendering the graphs. Also the -preview command my allow you to have a look at an approximate DAG without having to run the pipeline.
If you just try to install your dependencies using conda or mamba, there's no guarantee that the dependency graph will be solved the same way forever, therefore there is a chance your programs will work slightly different if you try to reinstall your dependencies sometime in the future.
That is why the golden rule for dependencies is to use a program like conda or mamba to intall them, but to do so inside a container image.
The container images are supposed to work the same way forever so doing so in this way each programs should install the same dependencies and produce the exact same enviroment for better reproducibility.
E.g.
env.yml
name: nf-tutorial
channels:
- conda-forge
- defaults
- bioconda
dependencies:
- bioconda::salmon=1.5.1
- bioconda::fastqc=0.11.9
- bioconda::multiqc=1.12
- conda-forge::tbb=2020.2Dockerfile
FROM mambaorg/micromamba:0.25.1
LABEL image.author.name "Your Name Here"
LABEL image.author.email "your@email.here"
COPY --chown=$MAMBA_USER:$MAMBA_USER env.yml /tmp/env.yml
RUN micromamba create -n nf-tutorial
RUN micromamba install -y -n nf-tutorial -f /tmp/env.yml && \
micromamba clean --all --yes
ENV PATH /opt/conda/envs/nf-tutorial/bin:$PATH
In your nextflow.config file, you can specifiy which container you want your Nextflow script file to use using the params.container parameter.
Example
params.container = 'nextflow/rnaseq-nf'
docker.runOptions = '-u $(id -u):$(id -g)'Because you are not providing any container repository or container registry for the script to use, by default Nextflow will try to pull the container from Docker Hub.
However, it will only try to pull the container if you tell Nextflow to run the pipeline with containers. If you want to run it with docker for example, you can run the script as follows:
nextflow run main.nf -with-dockerEach program should have its own designated container. Don't create container images with too many things or things your don't need.
Run only one process per container: In almost all cases, you should only run a single process in a single container. Decoupling applications into multiple containers makes it much easier to scale horizontally and reuse containers. If that service depends on another service, make use of container linking.
Biocontainers is a project to create a docker container for every recipe (package and version) they have in bioconda.
Sometimes you'll need to have a container with more than one tool, in this case there is a mulled container.
You can request a multi-package container here: https://biocontainers.pro/multipackage
If a package that you need is missing from Biocontainers, you'll likely have to create your own container as shown here.
You can also install galaxy-util-tools and search for mulled containers in your CLI using the mulled-search tool that comes with this tool
conda activate a-conda-env-you-already-have
conda install galaxy-tool-util
mulled-search --destination quay singularity --channel bioconda --search bowtie samtools | grep mulledAn even better solution than using Biocontainers is to use the Seqera labs Wave containers service. Wave is an open source software and a service that builds containers on the fly for you. You can just give your conda directives on Nextflow on a conda environment file and remotely Wave is going to build your container image on the fly during the execution of your pipeline and provide it back to you very quickly.
You can see more about what Wave containers can do with the following resrouces:
Wave containers containing conda packages can be built without the need for a Dockerfile. To do this, perfrom the following steps:
- In the
nextflow.configfile, add:
docker {
enabled = true
runOptions = '-u $(id -u):$(id -g)'
}
wave {
strategy = 'conda'
}- In your
processblock, add acondadirective. Example:
process SUBSET_FASTQ {
conda 'seqtk'
input:
tuple val(meta), path(fastqFile)
output:
tuple val(meta), path("${params.subsetCount}_rand-samples-from_${fastqFile}")
script:
"""
seqtk sample ${fastqFile} ${params.subsetCount} | gzip > ${params.subsetCount}_rand-samples-from_${fastqFile}
"""
}- Run the Nextflow pipeline with the
-with-waveoption:
nextflow run main.nf -with-waveNow Nextflow will automatically build a container containing the conda package that you requested.
Wavelit is the CLI tool for working with Wave outside of Nextflow. It allows you to use all the Wave functionality straight from the terminal.
You can find more information on Wavelit with the following resources:
Seqera Containers take the experience of Wave one step further. Instead of browsing available images as you would with a traditional container registry, you just type in the names of the tools you want to use into the following link https://seqera.io/containers/. Clicking “Get container” returns a container URI instantly, which you can use for anything - Nextflow pipeline or not. The key difference with Seqera Containers is that the image is also stored in an image cache, with infrastructure provided by AWS. Subsequent requests for the same package set will return the same image, ensuring reproducibility across runs. The cache has no expiry date, so those images will still be there if you need to rerun your analysis in the future (for at least 5 years after creation).
Seqera containers can be used directly from Nextflow and not only through https://seqera.io/containers/.
In order to use Seqera Containers in Nextflow, simply set wave.freeze without setting wave.build.repository - for example, by using the following config for your pipeline:
wave.enabled = true
wave.freeze = true
wave.strategy = 'conda'Any processes in your pipeline specifying Conda packages will have Docker or Singularity images created on the fly (depending on whether singularity.enabled is set or not) and cached for immediate access in subsequent runs. These images will be publicly available. You can view all container image names with the nextflow inspect command.
The file() method returns a Path, so any method defined for Path can also be used in a Nextflow script.
Additionally, the following methods are also defined for Paths in Nextflow:
-
exists()Returns true if the file exists. -
getBaseName()Gets the file name without its extension, e.g./some/path/file.tar.gz->file.tar. -
getExtension()Gets the file extension, e.g./some/path/file.txt->txt. -
getName()Gets the file name, e.g./some/path/file.txt->file.txt. -
getSimpleName()Gets the file name without any extension, e.g./some/path/file.tar.gz->file. -
getParent()Gets the file parent path, e.g./some/path/file.txt->/some/path. -
isDirectory()Returns true if the file is a directory. -
isEmpty()Returns true if the file is empty or does not exist. -
isFile()Returns true if it is a regular file (i.e. not a directory). -
isLink()Returns true if the file is a symbolic link.
Note
The getBaseName method drops the first file extension. If a file has more than one extension, for example reads.fastq.gz, calling getBaseName on this object will return reads.fastq. Since getBaseName acts on a sun.nio.fs.UnixPath object but returns a java.lang.String object, it can not be chained together like methods that act on and return file path objects like getParent.
To drop multiple extensions from the file name and get just the file name reads, add the implicit function file() between chains of getBaseName.
For example:
def filePath = file('reads.fastq.gz')
// INCORRECT
def fileNameOnly = filePath.getBaseName().getBaseName()
/* Outputs:
Exception thrown
groovy.lang.MissingMethodException: No signature of method: java.lang.String.getBaseName() is applicable for argument types: () values: []
*/
// CORRECT
def fileNameOnly = file(filePath.getBaseName()).getBaseName()
println(fileNameOnly)
// Outputs: readsProcess directives are optional settings that affect the execution of the process. They are written at the top of a process block.
Example
/*
* Process 1C: Create the genome index file for STAR
*/
process prepare_star_genome_index {
container 'quay.io/biocontainers/star:2.7.10b--h6b7c446_1'
input:
path genome
output:
path 'genome_dir'
script:
"""
mkdir -p genome_dir
STAR --runMode genomeGenerate \
--genomeDir genome_dir \
--genomeFastaFiles ${genome} \
--runThreadN ${task.cpus}
"""
}The container process directive tells Nextflow that if it is using docker, then to use that specific container for this specific task.
Process directives can be accessed for a specific task using the task. implicit variable. In the context of Nextflow, an implicit variable refers to a special type of variable that holds information about the execution context of a process.
Process directives can also be defaults and invisible. For example. The default number of CPUs to run a task is 1. You can pass this into a process parameter by using tasks.cpus. If you want to change this however, you can write at the top of your process block
cpus = 4The debug directive echos out the stdout from each of the tasks into the Nextflow stdout. By default the stdout produced by the commands executed in all processes is ignored.
Example:
process sayHello {
debug true
script:
"echo Hello"
}Outputs (in the shell terminal):
Hello
Whenever you have multiple samples being processed in a process, it's useful to use the .tag process directive.
The .tag process directive doesn't change anything in the analysis, but it allows you to associate each process execution with a custom label to make it easier to identify them in the log file or the trace execution report. This is useful if you want to know more information about multiple samples that are being run (which ones passed and which ones failed).
Example
/*
* Process 3: GATK Split on N
*/
process rnaseq_gatk_splitNcigar {
container 'quay.io/broadinstitute/gotc-prod-gatk:1.0.0-4.1.8.0-1626439571'
tag "${replicateId}"
input:
path genome
path index
path genome_dict
tuple val(replicateId), path(bam), path(bai)
output:
tuple val(replicateId), path('split.bam'), path('split.bai')
script:
"""
java -jar /usr/gitc/GATK35.jar -T SplitNCigarReads \
-R ${genome} -I ${bam} \
-o split.bam \
-rf ReassignOneMappingQuality \
-RMQF 255 -RMQT 60 \
-U ALLOW_N_CIGAR_READS \
--fix_misencoded_quality_scores
"""
}Will give you detials on every replicate ID that is being processed.
As of December 2023, Nextflow does not allow for keyword arguments. Therefore if you're trying to for example pass in 2 inputs, you look at the process block and the first input will be the first positional argument and the second input will be the second positional argument (and so on and so forth if there are more inputs)
Example
/*
* Process 1D: Create a file containing the filtered and recoded set of variants
*/
process prepare_vcf_file {
container 'quay.io/biocontainers/mulled-v2-b9358559e3ae3b9d7d8dbf1f401ae1fcaf757de3:ac05763cf181a5070c2fdb9bb5461f8d08f7b93b-0'
input:
path variantsFile
path blacklisted
output:
tuple path("${variantsFile.baseName}.filtered.recode.vcf.gz"),
path("${variantsFile.baseName}.filtered.recode.vcf.gz.tbi")
script:
"""
vcftools --gzvcf ${variantsFile} -c \
--exclude-bed ${blacklisted} \
--recode | bgzip -c \
> ${variantsFile.baseName}.filtered.recode.vcf.gz
tabix ${variantsFile.baseName}.filtered.recode.vcf.gz
"""
}
workflow {
reads_ch = Channel.fromFilePairs(params.reads)
prepare_genome_samtools(params.genome)
prepare_genome_picard(params.genome)
prepare_star_genome_index(params.genome)
// CORRECT
prepare_vcf_file(params.variants, params.blacklist)
// INCORRECT
prepare_vcf_file(variantsFile = params.variants, blacklisted = params.blacklist)
}Example
/*
* Process 4: GATK Recalibrate
*/
process rnaseq_gatk_recalibrate {
container 'quay.io/biocontainers/mulled-v2-aa1d7bddaee5eb6c4cbab18f8a072e3ea7ec3969:f963c36fd770e89d267eeaa27cad95c1c3dbe660-0'
tag "${replicateId}"
input:
path genome
path index
path dict
tuple val(replicateId), path(bam), path(bai)
tuple path(prepared_variants_file),
path(prepared_variants_file_index)
output:
tuple val(sampleId),
path("${replicateId}.final.uniq.bam"),
path("${replicateId}.final.uniq.bam.bai")
script:
// Notice how the sampleID is modified with a regular expression before the script is executed
sampleId = replicateId.replaceAll(/[12]$/,'')
"""
gatk3 -T BaseRecalibrator \
--default_platform illumina \
-cov ReadGroupCovariate \
-cov QualityScoreCovariate \
-cov CycleCovariate \
-knownSites ${prepared_variants_file} \
-cov ContextCovariate \
-R ${genome} -I ${bam} \
--downsampling_type NONE \
-nct ${task.cpus} \
-o final.rnaseq.grp
gatk3 -T PrintReads \
-R ${genome} -I ${bam} \
-BQSR final.rnaseq.grp \
-nct ${task.cpus} \
-o final.bam
(samtools view -H final.bam; samtools view final.bam | \
grep -w 'NH:i:1') | samtools view -Sb - > ${replicateId}.final.uniq.bam
samtools index ${replicateId}.final.uniq.bam
"""
}You can place the return keyword in your workflow block to aburptly stop the run at a certain point. This is really useful if you're troubleshooting and you just want to get the details up to a certain part of your workflow, the rest of it will remain untouched.
Example
workflow {
reads_ch = Channel.fromFilePairs(params.reads)
// The return keyword helps you troubleshoot.
// Here you want to check what the read ares
// The return keyword ensures the rest of the workflow doesn't get run
reads.view()
return
prepare_genome_samtools(params.genome)
prepare_genome_picard(params.genome)
prepare_star_genome_index(params.genome)
prepare_vcf_file(params.variants, params.blacklist)
rnaseq_mapping_star(params.genome, prepare_star_genome_index.out, reads_ch)
rnaseq_gatk_splitNcigar(params.genome,
prepare_genome_samtools.out,
prepare_genome_picard.out,
rnaseq_mapping_star.out)
rnaseq_gatk_recalibrate(params.genome,
prepare_genome_samtools.out,
prepare_genome_picard.out,
rnaseq_gatk_splitNcigar.out,
prepare_vcf_file.out)
// New channel to aggregate bam from different replicates into sample level.
rnaseq_gatk_recalibrate.out
| groupTuple
| set { recalibrated_samples }
rnaseq_call_variants(params.genome,
prepare_genome_samtools.out,
prepare_genome_picard.out,
recalibrated_samples)
post_process_vcf(rnaseq_call_variants.out,
prepare_vcf_file.out)
prepare_vcf_for_ase(post_process_vcf.out)
recalibrated_samples
.join(prepare_vcf_for_ase.out.vcf_for_ASE)
.map { meta, bams, bais, vcf -> [meta, vcf, bams, bais] }
.set { grouped_vcf_bam_bai_ch }
ASE_knownSNPs(params.genome,
prepare_genome_samtools.out,
prepare_genome_picard.out,
grouped_vcf_bam_bai_ch)
}You can also use this simple one liner that employs ; to be treated as a multiline expression which is very useful for debugging
| view { it }; return; //In vscode you can make use this code to make this a custom user snippet
{
"Nextflow debug snippet": {
"scope": "nextflow,nf,groovy",
"prefix": "vv",
"body": [
"| view { it }; return; //"
],
"description": "Inserts | view { it }; return; // for debugging"
}
}Because this returns the view command with a closure, you can easily modify input of the view command by modifying it. For example:
| view { it.class }; return; //^ This will tell you what the class is of the elements in your workflow.
Here are some more user snippets that are alterations of the snippet above.
{
"Nextflow debug snippet - basic": {
"scope": "nextflow,nf,groovy",
"prefix": "vv",
"body": [
"| view { it }; return; //"
],
"description": "Inserts | view { it }; return; // for debugging"
},
"Nextflow debug snippet - with count": {
"scope": "nextflow,nf,groovy",
"prefix": "cc",
"body": [
"| count | view { it }; return; //"
],
"description": "Inserts | count | view { it }; return; // for debugging"
},
"Nextflow debug snippet - detailed": {
"scope": "nextflow,nf,groovy",
"prefix": "vvv",
"body": [
"| view { println(it.class); println(it.size()); it }; return; //"
],
"description": "Inserts | view { println(it.class); println(it.size()); it }; return; // for debugging"
},
"Nextflow debug snippet - Hide Metamap": {
"scope": "nextflow,nf,groovy",
"prefix": "hmm",
"body": [
"| map { it -> [\"[ -- List of ${it[0].size()} metadata elements -- ]\", it[1]] } // HIDE METAMAP"
],
"description": "Replaces the metamap with an empty list to declutter the output"
},
"Nextflow debug snippet - Hide FilePath": {
"scope": "nextflow,nf,groovy",
"prefix": "hff",
"body": [
"| map { it -> [it[0], \"[ -- List of ${it[1].size()} files -- ]\"] } // HIDE FILEPATHS"
],
"description": "Replaces the file path with an empty string to declutter the output"
},
"Nextflow debug snippet - Hide Full FilePath": {
"scope": "nextflow,nf,groovy",
"prefix": "hfff",
"body": [
"| map { it -> [it[0], it[1].collect { filepath -> filepath.getName() }] } // HIDE FULL FILE PATHS"
],
"description": "Hides the full path name of the file object, giving you only just the basename to declutter the output"
},
}A common input pattern you see on a lot of Nextflow pipelines is
input:
tuple val(meta), path(files)Which means that the input is comprised of a tuple containing metadata and a path to files. Specifically, val(meta) is a placeholder for any type of metadata you want to associate with the input files, such as sample IDs, conditions, or other relevant information. The path(files) indicates the location of the actual data files that will be processed by the pipeline.
An example is taken from the nf-core/bacass pipelines below. Take a look at the output to see what the tuple val(meta), path(files) pattern looks like in practice.
ch_trim_reads
.join(ch_trim_json)
.map {
meta, reads, json ->
if (getFastpReadsAfterFiltering(json) > 0) {
[ meta, reads ]
}
}
.map { println it }
.set { ch_trim_reads }
// Outputs
// [[id:ERR064912, gsize:2.8m, single_end:false], [/Users/markpampuch/Dropbox/to_learn/nextflow/test-bacass/work/88/a5045082c5d077bd6fff234bea8388/ERR064912_1.fastp.fastq.gz, /Users/markpampuch/Dropbox/to_learn/nextflow/test-bacass/work/88/a5045082c5d077bd6fff234bea8388/ERR064912_2.fastp.fastq.gz]]This is what a proper standard input should look like. A list of metadata (a metamap) followed by a list of paths to your data files.
You can modify this standard input object slightly with maps to inspect the individual elements further.
Inspecting the metadata:
ch_trim_reads
.join(ch_trim_json)
.map {
meta, reads, json ->
if (getFastpReadsAfterFiltering(json) > 0) {
[ meta, reads ]
}
}
.map { it -> ["[ -- List of ${it[0].size()} metadata elements -- ]", it[1]] } // HIDE METAMAP
.map { println it }
.set { ch_trim_reads }
// Outputs
// [[ -- List of 3 metadata elements -- ], [/Users/markpampuch/Dropbox/to_learn/nextflow/test-bacass/work/88/a5045082c5d077bd6fff234bea8388/ERR064912_1.fastp.fastq.gz, /Users/markpampuch/Dropbox/to_learn/nextflow/test-bacass/work/88/a5045082c5d077bd6fff234bea8388/ERR064912_2.fastp.fastq.gz]]Inspecting the file paths:
ch_trim_reads
.join(ch_trim_json)
.map {
meta, reads, json ->
if (getFastpReadsAfterFiltering(json) > 0) {
[ meta, reads ]
}
}
.map { it -> ["[ -- List of ${it[0].size()} metadata elements -- ]", it[1]] } // HIDE METAMAP
.map { it -> [it[0], it[1].collect { filepath -> filepath.getName() }] } // HIDE FULL FILE PATHS
.map { println it }
.set { ch_trim_reads }
// Outputs
// [[ -- List of 3 metadata elements -- ], [ERR064912_1.fastp.fastq.gz, ERR064912_2.fastp.fastq.gz]]Inspecting the file paths further:
ch_trim_reads
.join(ch_trim_json)
.map {
meta, reads, json ->
if (getFastpReadsAfterFiltering(json) > 0) {
[ meta, reads ]
}
}
.map { it -> ["[ -- List of ${it[0].size()} metadata elements -- ]", it[1]] } // HIDE METAMAP
.map { it -> [it[0], it[1].collect { filepath -> filepath.getName() }] } // HIDE FULL FILE PATHS
.map { it -> [it[0], "[ -- List of ${it[1].size()} files -- ]"] } // HIDE FILEPATHS
.map { println it }
.set { ch_trim_reads }
// Outputs
// [[ -- List of 3 metadata elements -- ], [ -- List of 2 files -- ]]Inside a Nextflow workflow repository there are a couple special directories that are treated differently to other directories. These are the ./bin, ./templates, and the ./lib directory.
The bin directory (if it exists) is always added to the $PATH for all tasks. If the tasks are performed on a remote machine, the directory is copied across to the new machine before the task begins. This Nextflow feature is designed to make it easy to include accessory scripts directly in the workflow without having to commit those scripts into the container. This feature also ensures that the scripts used inside of the workflow move on the same revision schedule as the workflow itself.
It is important to know that Nextflow will take care of updating $PATH and ensuring the files are available wherever the task is running, but will not change the permissions of any files in that directory. If a file is called by a task as an executable, the workflow developer must ensure that the file has the correct permissions to be executed.
For example, let's say you have a small R script that produces a csv and a tsv:
#!/usr/bin/env Rscript
library(tidyverse)
plot <- ggplot(mpg, aes(displ, hwy, colour = class)) + geom_point()
mtcars |> write_tsv("cars.tsv")
ggsave("cars.png", plot = plot)You'd like to use this script in a simple workflow:
process PlotCars {
container 'rocker/tidyverse:latest'
output:
path("*.png"), emit: "plot"
path("*.tsv"), emit: "table"
script:
"""
cars.R
"""
}
workflow {
PlotCars()
PlotCars.out.table | view { "Found a tsv: $it" }
PlotCars.out.plot | view { "Found a png: $it" }
}Note
If you take a look in the R script, you'll notice that the tidyverse package was loaded using library(tidyverse). In order for this to work, the tidyverse package has to be somewhere where the R script could access it. This is why in the process block calling the R script, there is the process directive container 'rocker/tidyverse:latest' that loads in a container containing tidyverse.
To do this, you can create the bin directory, write our R script into the directory. Finally, and crucially, make the script executable:
mkdir -p bin
cat << EOF > bin/cars.R
#!/usr/bin/env Rscript
library(tidyverse)
plot <- ggplot(mpg, aes(displ, hwy, colour = class)) + geom_point()
mtcars |> write_tsv("cars.tsv")
ggsave("cars.png", plot = plot)
EOF
chmod +x bin/cars.RLet's run the script and see what Nextflow is doing for us behind the scenes:
cat << EOF > Nextflow.config
profiles {
docker {
docker.enabled = true
}
}
EOF
rm -r work
nextflow run . -profile dockerand then inspect the .command.run file that Nextflow has generated
code work/*/*/.command.runYou'll notice a nxf_container_env bash function that appends our bin directory to $PATH:
nxf_container_env() {
cat << EOF
export PATH="\$PATH:/workspace/gitpod/nf-training/advanced/structure/bin"
EOF
}Note
When working on the cloud, this nxf_container_env function will be slightly different. Nextflow will first also ensure that the bin directory is copied onto the virtual machine running your task in addition to the modification of $PATH.
Note
Always use a portable shebang line in your bin directory scripts.
-
In the R script example shown above, the
Rscriptprogram may be installed at (for example)/opt/homebrew/bin/Rscript. If you hard-code this path intocars.R, everything will work when you're testing locally outside of the docker container, but will fail when running with docker/singularity or in the cloud as theRscriptprogram may be installed in a different location in those contexts. -
It is strongly recommended to use
#!/usr/bin/envwhen setting the shebang for scripts in thebindirectory to ensure maximum portability.
If a process script block is becoming too long, it can be moved to a template file. The template file can then be imported into the process script block using the template method. This is useful for keeping the process block tidy and readable. Nextflow's use of $ to indicate variables also allows for directly testing the template file by running it as a script.
For example, let's say you want to run a python script inside a Nextflow process. You can set up your process like this:
process SayHi {
debug true
input:
val(name)
script:
template 'adder.py'
}Then inside a file called ./templates/adder.py you can have the following code:
#!/usr/bin/env python3
print("Hello $name!")
print("Process completed")Note
Notice how this script is not quite python because it's using $ string interpolation. This is as if you had written the script block like this instead:
process SayHi {
debug true
input:
val(name)
script:
"""
#!/usr/bin/env python3
print("Hello $name!")
print("Process completed")
"""
}Caution
The ./lib directory was removed from the nf-core template as of v2.13 (see here for more information).
The information in this section may be outdated / no longer good practice.
It may at times be helpful to bundle functionality into a new Groovy class (e.g. if a Groovy function that you need starts to grow with a lot of complexity). Any classes defined in the lib directory are added to the classpath and are available for use in the workflow - both main.nf and any imported modules.
Classes defined in lib directory can be used for a variety of purposes. For example, the nf-core/rnaseq workflow uses five custom classes:
NfcoreSchema.groovyfor parsing the schema.json file and validating the workflow parameters.NfcoreTemplate.groovyfor email templating and nf-core utility functions.Utils.groovyfor provision of a singlecheckCondaChannelsmethod.WorkflowMain.groovyfor workflow setup and to call theNfcoreTemplateclass.WorkflowRnaseq.groovyfor the workflow-specific functions.
The classes listed above all provide utility executed at the beginning of a workflow, and are generally used to "set up" the workflow. However, classes defined in lib can also be used to provide functionality to the workflow itself.
Let's consider an example where we create our own custom class to handle metadata. We can create a new class in ./lib/Metadata.groovy. We'll extend the built-in HashMap class, and add a simple method to return a value:
class Metadata extends HashMap {
def hi() {
return "Hello, workshop participants!"
}
}We can then use the new hi method in the workflow:
workflow {
Channel.of("Montreal")
| map { new Metadata() }
| view { it.hi() }
}At the moment, the Metadata class is not making use of the "Montreal" being passed into the closure. You can change that by adding a constructor to the class:
class Metadata extends HashMap {
Metadata(String location) {
this.location = location
}
def hi() {
return this.location ? "Hello, from ${this.location}!" : "Hello, workshop participants!"
}
}Which we can use like so:
workflow {
Channel.of("Montreal")
| map { new Metadata(it) }
| view { it.hi() }
}We can also use this method when passing the object to a process:
process UseMeta {
input: val(meta)
output: path("out.txt")
script: "echo '${meta.hi()}' | tee out.txt"
}
workflow {
Channel.of("Montreal")
| map { place -> new Metadata(place) }
| UseMeta
| view
}This might be helpful because you can add extra classes to the metadata which can be computed from the existing metadata. For example, we might want want to grab the adapter prefix:
def getAdapterStart() {
this.adapter?.substring(0, 3)
}Which we might use like so:
process UseMeta {
input: val(meta)
output: path("out.txt")
script: "echo '${meta.adapter} prefix is ${meta.getAdapterStart()}' | tee out.txt"
}
workflow {
Channel.of("Montreal")
| map { place -> new Metadata(place) }
| map { it + [adapter:"AACGTAGCTTGAC"] }
| UseMeta
| view
}You might even want to reach out to external services such as a LIMS or the E-utilities API. Here we add a dummy "getSampleName()" method that reaches out to a public API:
import groovy.json.JsonSlurper
// ...
def getSampleName() {
def get = new URL('https://postman-echo.com/get?sampleName=Fido').openConnection()
def getRC = get.getResponseCode();
if (getRC.equals(200)) {
JsonSlurper jsonSlurper = new JsonSlurper()
def json = jsonSlurper.parseText(get.getInputStream().getText())
return json.args.sampleName
}
}Which we can use like so:
process UseMeta {
input: val(meta)
output: path("out.txt")
script:
"echo '${meta.adapter} prefix is ${meta.getAdapterStart()} with sampleName ${meta.getSampleName()}' | tee out.txt"
}Note
When we start passing custom classes through the workflow, it's important to understand a little about the Nextflow caching mechanism. When a task is run, a unique hash is calculated based on the task name, the input files/values, and the input parameters. Our class extends from HashMap, which means that the hash will be calculated based on the contents of the HashMap. If we add a new method to the class, or amend a class method, this does not change the value of the objects in the hash, which means that the hash will not change. Example:
- We might increase the length of the adapter prefix to 5 characters:
def getAdapterStart() {
this.adapter?.substring(0, 5)
}- Changing this method and resuming the workflow will not change the hash, and the existing method will be used.
We are not limited to using or extending the built-in Groovy classes. For example, if you have created a Dog class in ./lib/Dog.groovy:
class Dog {
String name
Boolean isHungry = true
}We can create a new dog at the beginning of the workflow:
workflow {
dog = new Dog(name: "fido")
log.info "Found a new dog: $dog"
}We can pass objects of our class through channels. Here we take a channel of dog names and create a channel of dogs:
workflow {
Channel.of("Argente", "Absolon", "Chowne")
| map { new Dog(name: it) }
| view
}If we try to use this new class in a resumed process, no caches will be used.
Nextflow has provided a decorator to help serialize your custom classes. By adding @ValueObject to the class definition, Nextflow will automatically serialize the class and cache it. This is useful if you want to pass a custom class through a channel, or if you want to use the class in a resumed workflow.
To do this, we first add the decorator to the desired class:
import Nextflow.io.ValueObject
@ValueObject
class Dog {
String name
Boolean isHungry = true
}By adding the @ValueObject decorator to the Dog class, Nextflow will now use the properties name and isHungry as the elements that are be hashed to provide the input to the task and to check if the task outputs can be pulled from the cache.
Lastly, we will need to register the class with Kryo, the Java serialization framework. Again, Nextflow provides a helper method to do this. We can add the following to the main.nf file:
import Nextflow.util.KryoHelper
KryoHelper.register(Dog)Now the Dog class can now be used in processes and cached correctly.
Because Nextflow is a deeply asyncronous system, modifying data in place can affect the results of another process that may be trying to access the same memory. Whenever you need to modify data, make sure to perform operations that create a copy or new object for the data.
For example, this code intentionally performs data modifications in-place:
workflow {
Channel.fromPath("data/samplesheet.csv")
| splitCsv( header:true )
| map { row ->
meta = row.subMap('id', 'repeat', 'type')
[meta, [
file(row.fastq1, checkIfExists: true),
file(row.fastq2, checkIfExists: true)]]
}
| set { samples }
samples
| map { sleep 10; it }
| view { meta, reads -> "Should be unmodified: $meta" }
samples
| map { meta, reads ->
meta.type = "BROKEN"
[meta, reads]
}
| view { meta, reads -> "Should be MODIFIED: $meta" }
}What you would expect is to have outputs from two channels, one with the data as was (unmodified) and one with the data modified. This is what you get as output instead.
Should be MODIFIED: [id:sampleA, repeat:1, type: BROKEN]
Should be MODIFIED: [id:sampleA, repeat:1, type: BROKEN]
Should be MODIFIED: [id:sampleA, repeat:2, type:BROKEN]
Should be MODIFIED: [id:sampleA, repeat:2, type: BROKEN]
Should be MODIFIED: [id:sampleB, repeat:1, type: BROKEN]
Should be MODIFIED: [id:sampleB, repeat:1, type:BROKEN]
Should be MODIFIED: [id:sampleC, repeat:1, type: BROKEN]
Should be MODIFIED: [id:sampleC, repeat:1, type: BROKEN]
Should be unmodified: [id:sampleA, repeat:1, type: BROKEN]
Should be unmodified: [id:sampleA, repeat:1, type: BROKEN]
Should be unmodified: [id:sampleA, repeat:2, type: BROKEN]
Should be unmodified: [id:sampleA, repeat:2, type: BROKEN]
Should be unmodified: [id:sampleB, repeat:1, type: BROKEN]
Should be unmodified: [id:sampleB, repeat:1, type: BROKEN]
Should be unmodified: [id:sampleC, repeat:1, type: BROKEN]
Should be unmodified: [id:sampleC, repeat:1, type: BROKEN]
Instead you have all the data having been modified because both maps were performing over the same meta object in memory.
To make this operation safe, you must ensure that you are using operators that return copies (e.g. subMap, +, etc.)
Example:
workflow {
Channel.fromPath("data/samplesheet.csv")
| splitCsv( header:true )
| map { row ->
meta = row.subMap('id', 'repeat', 'type')
[meta, [
file(row.fastq1, checkIfExists: true),
file(row.fastq2, checkIfExists: true)]]
}
| set { samples }
samples
| map { sleep 10; it }
| view { meta, reads -> "Should be unmodified: $meta" }
samples
| map { meta, reads ->
[meta + [type: "BROKEN"], reads]
}
| view { meta, reads -> "Should be MODIFIED: $meta" }
}Outputs:
Should be MODIFIED: [id:sampleA, repeat:1, type: BROKEN]
Should be MODIFIED: [id:sampleA, repeat:1, type: BROKEN]
Should be MODIFIED: [id:sampleA, repeat:2, type:BROKEN]
Should be MODIFIED: [id:sampleA, repeat:2, type: BROKEN]
Should be MODIFIED: [id:sampleB, repeat:1, type: BROKEN]
Should be MODIFIED: [id:sampleB, repeat:1, type:BROKEN]
Should be MODIFIED: [id:sampleC, repeat:1, type: BROKEN]
Should be MODIFIED: [id:sampleC, repeat:1, type: BROKEN]
Should be unmodified: [id:sampleA, repeat:1, type: normal]
Should be unmodified: [id:sampleA, repeat:1, type: tumor]
Should be unmodified: [id:sampleA, repeat:2, type: normal]
Should be unmodified: [id:sampleA, repeat:2, type: tumor]
Should be unmodified: [id:sampleB, repeat:1, type: normal]
Should be unmodified: [id:sampleB, repeat:1, type: tumor]
Should be unmodified: [id:sampleC, repeat:1, type: normal]
Should be unmodified: [id:sampleC, repeat:1, type: tumor]
In Nextflow, it's recommended to keep all your flowing through your pipeline for as long as possible, even if you don't need it. There's very little cost to holding onto your metadata and keeping if flowing through the directed acyclic graph. You may never know when you'll need that data and once it becomes discociated from the grouped data it's very hard to join it back up again.
An extremely common channel "shape" or cardinality in nf-core modules and subworkflows is the tuple val(meta), path(reads) structure.
Example:
process EXAMPLE_PROCESS {
input:
tuple val(meta), path(reads)
// ...This style is critical to enabling reusability of these modules. If you structure your data to have a tuple containing a "metadata map" and the path to your data your data would be suitable for any Nextflow process with inputs in the above form.
Example:
workflow {
Channel.fromFilePairs("data/reads/*/*_R{1,2}.fastq.gz")
| map { id, reads ->
(sample, replicate, type) = id.tokenize("_")
(treatmentFwd, treatmentRev) = reads*.parent*.name*.minus(~/treatment/)
meta = [
sample:sample,
replicate:replicate,
type:type,
treatmentFwd:treatmentFwd,
treatmentRev:treatmentRev,
]
[meta, reads]
}
| view
}More information on metadata propogation can be found here.
Using grouping you can constuct more complicated graph structures inside your workflow by taking data from multiple processes, splitting it apart, and then putting it back together. See here for some of the best practices for doing so.
Most Nextflow users prefer the set notation rather than the = notation
Example:
workflow {
// Create a channel with all the fastq files in the data directory
Channel.fromPath("${params.dataDir}/**/*.fastq.gz")
| map { path ->
// Extract the part of the path that is to the right of params.dataDir
def relativePath = file(path.getParent().toString().split(params.dataDir)[1])
// Create a tuple with the relative path and the file path
Tuple.tuple(relativePath, path)
}
| set { fastqFiles_ch }
// Subset the fastq files
SUBSET_FASTQ(fastqFiles_ch)
| set { fastqFilesSubsetted_ch }
}Instead of:
workflow {
// Create a channel with all the fastq files in the data directory
fastqFiles_ch = Channel.fromPath("${params.dataDir}/**/*.fastq.gz")
| map { path ->
// Extract the part of the path that is to the right of params.dataDir
def relativePath = file(path.getParent().toString().split(params.dataDir)[1])
// Create a tuple with the relative path and the file path
Tuple.tuple(relativePath, path)
}
// Subset the fastq files
fastqFilesSubsetted_ch = SUBSET_FASTQ(fastqFiles_ch)
fastqFilesSubsetted_ch.view()
}It’s good practice to organize each experiment in its own folder. The main experiment input parameters should be specified using a Nextflow config file. This makes it simple to track and replicate an experiment over time.
A work directory will be created inside of each experiment directory when the Nextflow script is launched from that directory.
In the same experiment, the same workflow can be executed multiple times, however, launching two (or more) Nextflow instances in the same directory concurrently should be avoided.
The nextflow log command lists the executions run in the current folder:
Ouputs:
TIMESTAMP DURATION RUN NAME STATUS REVISION ID SESSION ID COMMAND
2019-05-06 12:07:32 1.2s focused_carson ERR a9012339ce 7363b3f0-09ac-495b-a947-28cf430d0b85 Nextflow run hello
2019-05-06 12:08:33 21.1s mighty_boyd OK a9012339ce 7363b3f0-09ac-495b-a947-28cf430d0b85 Nextflow run rnaseq-nf -with-docker
2019-05-06 12:31:15 1.2s insane_celsius ERR b9aefc67b4 4dc656d2-c410-44c8-bc32-7dd0ea87bebf Nextflow run rnaseq-nf
2019-05-06 12:31:24 17s stupefied_euclid OK b9aefc67b4 4dc656d2-c410-44c8-bc32-7dd0ea87bebf Nextflow run rnaseq-nf -resume -with-docker
You can use either the session ID or the run name to recover a specific execution:
nextflow run rnaseq-nf -resume mighty_boydMore information can be found here.
Most times it's much easier to build a pipeline with a subset of your data. Here are some tips for how I would structure your data for building a pipeline.
- Copy all of your data into your Nextflow project and give it the directory name
data. - In a
README.mdfile, provide a path for where a backup of all the data can be found in case something goes wrong.- (usually all the data will be really big so copying it all in the directory for backup will waste a lot of space).
- Create a subset of your data into your Nextflow project and give it the directory name
data_for-pipeline-dev.- Take your time with this. The development data architecture should exactly match the architecutre of the data in the
datafolder. This development data should be like if you just shrunk the data into the minimum usable data to get the processes in your pipeline working.
- Take your time with this. The development data architecture should exactly match the architecutre of the data in the
- Create a copy of your subsetted data and call it
data_for-pipeline-dev_BACKUP. Don't touch this file. Copy it back todata_for-pipeline-devif you mess up your data. - Inside your
nextflow.configfile, add the following code:
params.devMode = true
params.dataDir = params.devMode ? 'data_for-pipeline-dev' : 'data'- (Optional) If you wish to further modify your development data only, you can perform conditional execution in your workflow using
if/elseand theparams.devModeparameter. For example, take a look at the followingmain.nffile:
#!/usr/bin/env nextflow
include { SUBSET_FASTQ } from './modules/local/subset_fastq/main.nf'
workflow {
// Create a channel with all the file metadata
Channel.fromPath("${params.dataDir}/**/*.fastq.gz")
| map { path ->
def experimentName = path.getParent().getParent().getParent().getParent().getBaseName()
(date, strain, replicate, extractionMethod) = experimentName.tokenize('_')
def extras = experimentName.split('--').size() == 2 ? experimentName.split('--')[1] : null
if (extras) {extractionMethod = extractionMethod.split('--')[0]}
def readsPassOrFail = path.getParent().getBaseName() - 'fastq_'
def id = file(path.getBaseName()).getBaseName()
def meta = [
id: id,
date: date,
strain: strain,
replicate: replicate - 'rep-',
extractionMethod: extractionMethod,
extras: extras,
readsPassOrFail: readsPassOrFail
]
return [meta, path]
}
| set { fastqFiles_ch }
// Subset the fastq files if in dev mode
if (params.devMode) {
SUBSET_FASTQ(fastqFiles_ch)
| map { meta, fastqFile ->
def subsetted = true
def numReadsInSubset = fastqFile.getBaseName().split('_')[0]
meta += [subsetted: subsetted, numReadsInSubset: numReadsInSubset]
return [meta, fastqFile]
}
| set { fastqFilesPostSubsetting_ch }
}
else {
fastqFiles_ch
| map { meta, fastqFile ->
def subsetted = false
def numReadsInSubset = null
meta += [subsetted: subsetted, numReadsInSubset: numReadsInSubset]
return [meta, fastqFile]
}
| set { fastqFilesPostSubsetting_ch }
}
}- When you want to run your actual pipeline, turn off development mode with
--devMode false
nextflow run main.nf -with-wave -resume --devMode falseNote
For more information on conditional workflow execution, see here.
A Nextflow pipeline can be represented as a direct acyclic graph (DAG). The nodes in the graph represent the pipeline’s processes and operators, while the edges represent the data dependencies (i.e. channels) between them.
To render the workflow DAG, run your pipeline with the -with-dag option. You can use the -preview option with -with-dag to render the workflow DAG without executing any tasks.
By default, it creates a file named dag-<timestamp>.html with the workflow DAG rendered as a Mermaid diagram. For example:
nextflow run main.nf -with-dag -previewWill output something like dag-20240507-85496265.html.
To change the output file format, you can pass a filename after the with-dag option. The supported file types for DAG visualization are:
.dot: Creates a Graphviz DOT file.gexf: Creates a Graph Exchange XML file (Gephi) file.html: (default) Creates a HTML file with Mermaid diagram.mmd: Creates mermaid diagram file (compatible with markdown).pdf: Creates a Graphviz PDF file (Requires Graphviz to be installed).png: Creates a Graphviz PNG file (Requires Graphviz to be installed).svg: Creates a Graphviz SVG file (Requires Graphviz to be installed)
Warning
(As of April 2024) The nf-core package (v2.13.1) from bioconda may be incompatible with the graphviz package (v9.0.0) from conda-forge. Be aware of having these both in the same conda environment.
However, the default filename (dag-<timestamp>.html) is dynamically generated to generate a timestamp. If you naively change the filename in order to try to change the output file type, there's a good chance you'll hardcode the filename and lose the dynamic timestamp generation, in order to get around this, you can leverage the shell command date. Rename the file to be "dag-$(date +"%Y%m%d-%H%M%S").FILETYPE".
For example, to create a mermaid diagram on a simple workflow I wrote, you can use:
nextflow run main.nf -with-wave -with-dag "dag-$(date +"%Y%m%d-%H%M%S").mmd" -previewThis will output:
flowchart TB
subgraph " "
v0["Channel.fromPath"]
end
v2([SUBSET_FASTQ])
subgraph " "
v4["fastqFilesSubsetted_ch"]
end
v1(( ))
v3(( ))
v0 --> v1
v1 --> v2
v2 --> v3
v3 --> v4
Note
The small nodes in the graph that don't have any text or names by them represent map operations on the data (think of these like anonymous function operations. That's why they have no names).
When you don't know how a specific Nextflow function or element works, a really good resource is seeing how it was implemented in nf-core. The nf-core repository contains dozens of professional and expertly curated pipelines. By going to https://github.com/nf-core and typing into the search bar a specific function or operator, you can get tons and tons of examples of how it is supposed to be used.
Warning
Seqera Platform was previously known as Nextflow Tower. You may see this name thrown around trying to find information online about it.
Seqera Platform provides a useful tool to monitor your pipelines and work on teams. It offered through a web app and a CLI. To learn more about how to use it you can click here:
-
Web app:
-
CLI:
Also can watch this video to learn how to get set up and use Seqera platform.
The Nextflow VSCode extension gives developers tooling designed to make the nextflow development experience smoother. It provides syntax highlighting, and as of recently, warning/error messages, hover hints, code completion, code navigation, automatic formatting, symbol renaming, parameter schema checking, and DAG previews for workflows as well.
To learn more about the features of this extension, click here.
Bioinformaticians often run their Nextflow workflows on internal HPC clusters, typically using job schedulers like Slurm to manage tasks. However, as the need for scalability and flexibility grows, many are making the shift to public cloud platforms. In these scenarios, platforms like Google Cloud Platform (GCP), with services such as Cloud Storage and the Google Batch API, become key to enabling this transition. The process of adapting workflows from HPC to the cloud involves a series of essential steps that are applicable not just to GCP, but to other cloud providers as well, making it a valuable transformation blueprint for the broader bioinformatics community.
To learn more about this migration process, read this blogpost here.
nf-core is a community effort to collect a curated set of analysis workflows built using Nextflow. nf-core provides a standardised set of best practices, guidelines, and templates for building and sharing bioinformatics workflows.
Warning
Just because nf-core pipelines are built using Nextflow, there is still a steep learning curve to using nf-core effectively even if you can read and write Nextflow code.
For that reason nf-core deserves it's own dedicated chapter this Nextflow notes document.
Tip
Beyond just reviewing these notes, in order to use, contribute to, and get the benefits out of using nf-core pipelines and the infrastructure nf-core provies in your work, it's important to always stay up to date with what the nf-core community is doing (read the docs, ask questions in the nf-core slack, follow the nf-core bytesize talks, check the nf-core blog, etc.)
nf-core pipelines follow a set of best practices and standardised conventions. nf-core pipelines start from a common template and follow the same structure. Although you won’t need to edit code in the pipeline project directory, having a basic understanding of the project structure and some core terminology will help you understand how to configure its execution.
Nextflow DSL2 workflow are built up of subworkflows and modules that are stored as separate .nf files.
Most nf-core pipelines consist of a single workflow file (there are a few exceptions). This is the main <workflow>.nf file that is used to bring everything else together. Instead of having one large monolithic script, it is broken up into a combination of subworkflows and modules.
More information on configuring these pipelines can be found here.
The nf-core pipeline template is a working pipeline and comes pre-configured with two modules:
-
FastQC: A tool that performs quality control checks on raw sequence data coming from high throughput sequencing pipelines. It provides a modular set of analyses that can be used to give a quick impression of your data. -
MultiQC: A modular tool to aggregate results from bioinformatics analyses across many samples into a single report.
To add a module to an nf-core pipelines, following the instructions in the documentation found here.
Most of the nf-core modules will be used somewhere in a subworkflow. However, there is one exception. The MultiQC module will typically be used at the end of the workflow, aggregating all the data from the previous outputs. This means that it is typically used differently than other nf-core modules.
It's not easy to write hard and fast rules for how to incorporate modules into your nextflow pipeline, because each of them are slightly different. Below I've instead written out sort of a case study detailing how I got preset modules to work in a pipeline I was modifying and my thoughts and investigative process at the time.
Here I describe how I added the nf-coresamtools_depth module to my pipeline.
- The first thing I did was install the module of interest into my pipeline. This was done using the
nf-core modules installcommand
nf-core modules install samtools/depthNote
If this doesn't work, there may be an issue with the nf-core cache. Try clearing it by removing ~/.config/nfcore
-
Next I looked for the part of my nf-core pipeline where I wanted this module to be inserted. In this case it was in the
./subworkflows/local/convert_stats.nffile. -
The
convert_stats.nffile has a lot of import statements
include { CRUMBLE } from '../../modules/nf-core/crumble/main'
include { SAMTOOLS_VIEW as SAMTOOLS_CRAM } from '../../modules/nf-core/samtools/view/main'
include { SAMTOOLS_VIEW as SAMTOOLS_REINDEX } from '../../modules/nf-core/samtools/view/main'
include { SAMTOOLS_STATS } from '../../modules/nf-core/samtools/stats/main'
include { SAMTOOLS_FLAGSTAT } from '../../modules/nf-core/samtools/flagstat/main'
include { SAMTOOLS_IDXSTATS } from '../../modules/nf-core/samtools/idxstats/main'- I added the path to the
SAMTOOLS_DEPTHmodule (making sure to link themain.nffile inside that directory but without the.nfextension).
include { CRUMBLE } from '../../modules/nf-core/crumble/main'
include { SAMTOOLS_VIEW as SAMTOOLS_CRAM } from '../../modules/nf-core/samtools/view/main'
include { SAMTOOLS_VIEW as SAMTOOLS_REINDEX } from '../../modules/nf-core/samtools/view/main'
include { SAMTOOLS_STATS } from '../../modules/nf-core/samtools/stats/main'
include { SAMTOOLS_FLAGSTAT } from '../../modules/nf-core/samtools/flagstat/main'
include { SAMTOOLS_IDXSTATS } from '../../modules/nf-core/samtools/idxstats/main'
include { SAMTOOLS_DEPTH } from '../../modules/nf-core/samtools/depth/main'- Now that the
SAMTOOLS_DEPTHmodule was in my subworkflow, I took a look at themain.nffile inside theSAMTOOLS_DEPTHmodule (file path:./modules/nf-core/samtools/depth/main.nf) to see what inputs it took. Inside themain.nffile, you can see this:
process SAMTOOLS_DEPTH {
tag "$meta1.id"
label 'process_low'
conda "${moduleDir}/environment.yml"
container "${ workflow.containerEngine == 'singularity' && !task.ext.singularity_pull_docker_container ?
'https://depot.galaxyproject.org/singularity/samtools:1.21--h50ea8bc_0' :
'biocontainers/samtools:1.21--h50ea8bc_0' }"
input:
tuple val(meta1), path(bam)
tuple val(meta2), path(intervals)
output:
tuple val(meta1), path("*.tsv"), emit: tsv
path "versions.yml" , emit: versions
when:
task.ext.when == null || task.ext.when
script:
def args = task.ext.args ?: ''
def prefix = task.ext.prefix ?: "${meta1.id}"
def positions = intervals ? "-b ${intervals}" : ""
"""
samtools \\
depth \\
--threads ${task.cpus-1} \\
$args \\
$positions \\
-o ${prefix}.tsv \\
$bam
cat <<-END_VERSIONS > versions.yml
"${task.process}":
samtools: \$(echo \$(samtools --version 2>&1) | sed 's/^.*samtools //; s/Using.*\$//')
END_VERSIONS
"""
}- The key portion of this file is the
inputblock:
input:
tuple val(meta1), path(bam)
tuple val(meta2), path(intervals)- Here you can see that this module takes in 2 inputs.
- A tuple containing a list of metadata for the file and a
.bamfile. - A tuple containing a list of metadata for the file and a file containing interval information for the
samtools depthprogram.
- A tuple containing a list of metadata for the file and a
- Typically, each module will only require 1 mandatory file, and other inputs might be optional. It's a good idea to investigate the rest of the module code to see if you can verify whether the other inputs are optional.
- Inside the
scriptblock of the module, you can see that the module uses this chunk of code
- Inside the
def positions = intervals ? "-b ${intervals}" : ""
"""
samtools \\
depth \\
--threads ${task.cpus-1} \\
$args \\
$positions \\
-o ${prefix}.tsv \\
$bam
// ...(rest of code below)- This chunk of code indicates that the the
intervalsposition is probably optional. Thepositionsvariable is used in the part of the code that executes thesamtools depthprogram and is passed as a parameter. However, this variable is set conditionally and one of the options is to set it as an empty string, indicating that this paramter can be a real value or nothing.- The
?and:syntax indicates the tertiary operator in Groovy. This is akin to anif elsestatement. This linedef positions = intervals ? "-b ${intervals}" : ""can be interpretted as "If the variableinterestsexists and contains real (truthy) data, set the variablepositionsto the value of the string"-b (the value inside the interests variable)", otherwise, ifintervalscontains a falsey value, setpositionsto an empty string ("").
- The
- Since I don't want to pass in an intervals file to the
samtools depthprogram, this means that the only input I'm concerned with is thetuple val(meta1), path(bam)input.
- Now I went back into my subworkflows file investigate where I wanted to call this
SAMTOOLS_DEPTHprogram and how I could pass the inputs into the file. Back in the./subworkflows/local/convert_stats.nffile. The entire file looked like this:
//
// Convert BAM to CRAM, create index and calculate statistics
//
include { CRUMBLE } from '../../modules/nf-core/crumble/main'
include { SAMTOOLS_VIEW as SAMTOOLS_CRAM } from '../../modules/nf-core/samtools/view/main'
include { SAMTOOLS_VIEW as SAMTOOLS_REINDEX } from '../../modules/nf-core/samtools/view/main'
include { SAMTOOLS_STATS } from '../../modules/nf-core/samtools/stats/main'
include { SAMTOOLS_FLAGSTAT } from '../../modules/nf-core/samtools/flagstat/main'
include { SAMTOOLS_IDXSTATS } from '../../modules/nf-core/samtools/idxstats/main'
include { SAMTOOLS_DEPTH } from '../../modules/nf-core/samtools/depth/main'
workflow CONVERT_STATS {
take:
bam // channel: [ val(meta), /path/to/bam, /path/to/bai ]
fasta // channel: [ val(meta), /path/to/fasta ]
main:
ch_versions = Channel.empty()
// Split outfmt parameter into a list
def outfmt_options = params.outfmt.split(',').collect { it.trim() }
// (Optionally) Compress the quality scores of Illumina and PacBio CCS alignments
if ( params.compression == "crumble" ) {
bam
| branch {
meta, bam ->
run_crumble: meta.datatype == "hic" || meta.datatype == "illumina" || meta.datatype == "pacbio"
no_crumble: true
}
| set { ch_bams }
CRUMBLE ( ch_bams.run_crumble, [], [] )
ch_versions = ch_versions.mix( CRUMBLE.out.versions )
// Convert BAM to CRAM
CRUMBLE.out.bam
| mix( ch_bams.no_crumble )
| map { meta, bam -> [meta, bam, []] }
| set { ch_bams_for_conversion }
} else {
bam
| map { meta, bam -> [meta, bam, []] }
| set { ch_bams_for_conversion }
}
// (Optionally) convert to CRAM if it's specified in outfmt
ch_cram = Channel.empty()
ch_crai = Channel.empty()
if ("cram" in outfmt_options) {
SAMTOOLS_CRAM ( ch_bams_for_conversion, fasta, [] )
ch_versions = ch_versions.mix( SAMTOOLS_CRAM.out.versions.first() )
// Combine CRAM and CRAI into one channel
ch_cram = SAMTOOLS_CRAM.out.cram
ch_crai = SAMTOOLS_CRAM.out.crai
}
// Re-generate BAM index if BAM is in outfmt
def ch_data_for_stats
if ("cram" in outfmt_options) {
ch_data_for_stats = ch_cram.join( ch_crai )
} else {
ch_data_for_stats = ch_bams_for_conversion
}
ch_bam = Channel.empty()
ch_bai = Channel.empty()
if ("bam" in outfmt_options) {
// Re-generate BAM index
SAMTOOLS_REINDEX ( ch_bams_for_conversion, fasta, [] )
ch_versions = ch_versions.mix( SAMTOOLS_REINDEX.out.versions.first() )
// Set the BAM and BAI channels for emission
ch_bam = SAMTOOLS_REINDEX.out.bam
ch_bai = SAMTOOLS_REINDEX.out.bai
// If using BAM for stats, use the reindexed BAM
if ( !("cram" in outfmt_options) ) {
ch_data_for_stats = ch_bam.join ( ch_bai )
}
}
// Calculate statistics
SAMTOOLS_STATS ( ch_data_for_stats, fasta )
ch_versions = ch_versions.mix( SAMTOOLS_STATS.out.versions.first() )
// Calculate statistics based on flag values
SAMTOOLS_FLAGSTAT ( ch_data_for_stats )
ch_versions = ch_versions.mix( SAMTOOLS_FLAGSTAT.out.versions.first() )
// Calculate index statistics
SAMTOOLS_IDXSTATS ( ch_data_for_stats )
ch_versions = ch_versions.mix( SAMTOOLS_IDXSTATS.out.versions.first() )
emit:
bam = ch_bam // channel: [ val(meta), /path/to/bam ] (optional)
bai = ch_bai // channel: [ val(meta), /path/to/bai ] (optional)
cram = ch_cram // channel: [ val(meta), /path/to/cram ] (optional)
crai = ch_crai // channel: [ val(meta), /path/to/crai ] (optional)
stats = SAMTOOLS_STATS.out.stats // channel: [ val(meta), /path/to/stats ]
flagstat = SAMTOOLS_FLAGSTAT.out.flagstat // channel: [ val(meta), /path/to/flagstat ]
idxstats = SAMTOOLS_IDXSTATS.out.idxstats // channel: [ val(meta), /path/to/idxstats ]
versions = ch_versions // channel: [ versions.yml ]
}- These were the most important lines for figuring out how to add this module.
// ...(rest of code)
workflow CONVERT_STATS {
take:
bam // channel: [ val(meta), /path/to/bam, /path/to/bai ]
// ...(rest of code)
// Calculate statistics
SAMTOOLS_STATS ( ch_data_for_stats, fasta )
ch_versions = ch_versions.mix( SAMTOOLS_STATS.out.versions.first() )
// ...(rest of code)- Here, you can see 2 important things:
- The subworkflow takes in a
bamchannel as input. The contents of this channel are a tuple containing the metadata of the.bamfile, the path to the.bamfile, and a path to the.baifile, which is an indexed version of the.bamfile. - The line
SAMTOOLS_STATS(ch_data_for_stats, fasta)is a call to thesamtools statsprogram, which is structured very similar to thesamtools depthprogram that we're trying to add. You can see that the program takes in a channel containing the data to process, calledch_data_for_stats, as well as a channel containing the path to the.fastafile. If you look at the entire subworkflow code, you can see that thech_data_for_statschannel contains basically the same information as thebamchannel, but the data was processed to be fed nicely to the other programs. Just like thebamchannel, thisch_data_for_statschannel contains a list of metadata for the.bamfile, the path to the.bamfile, and a path to the indexed.baifile.- Since the
SAMTOOLS_DEPTHmodule only required the metadata and the path to the.bamfile, but not the indexed file, we can use the channel operatormapto modify the channel and format it properly for this program.- This can be done with something like this:
ch_data_for_stats.map { meta, input, index -> [meta, input] }
- This can be done with something like this:
- Since the program also takes an optional
intervalsinput, we can create a dummy channel that has bascially an empty value that we can pass into the program to satisfy it's 2 input requirements, but not effect the processing of our data.- This can be done with something like this:
ch_intervals = ch_data_for_stats.map { meta, input, index -> [meta, []] }
- This can be done with something like this:
- Since the
- The subworkflow takes in a
- Therefore, to add a call to the
SAMTOOLS_DEPTHprogram, you can insert the following code:
// Using samtools depth
ch_data_for_samtools_depth = ch_data_for_stats.map { meta, input, index -> [meta, input] }
ch_intervals = ch_data_for_stats.map { meta, input, index -> [meta, []] }
SAMTOOLS_DEPTH(ch_data_for_samtools_depth, ch_intervals)
ch_versions = ch_versions.mix(SAMTOOLS_DEPTH.out.versions.first())Note
The line ch_versions = ch_versions.mix(SAMTOOLS_DEPTH.out.versions.first()) a standard best practice is used to emit the exact version of the program used. All the standard nextflow modules will contain lines in their script blocks that emit the version and an output channel called versions that you can use to capture that version information and put into one output file at the end of the program.
- The final step in this workflow file was to emit the data from the
SAMTOOLS_DEPTHmodule. At the bottom of the subworkflow, you can see this code block:
emit:
bam = ch_bam // channel: [ val(meta), /path/to/bam ] (optional)
bai = ch_bai // channel: [ val(meta), /path/to/bai ] (optional)
cram = ch_cram // channel: [ val(meta), /path/to/cram ] (optional)
crai = ch_crai // channel: [ val(meta), /path/to/crai ] (optional)
stats = SAMTOOLS_STATS.out.stats // channel: [ val(meta), /path/to/stats ]
flagstat = SAMTOOLS_FLAGSTAT.out.flagstat // channel: [ val(meta), /path/to/flagstat ]
idxstats = SAMTOOLS_IDXSTATS.out.idxstats // channel: [ val(meta), /path/to/idxstats ]
versions = ch_versions // channel: [ versions.yml ]
}- This code will emit the data that gets outputted by each module, so that whatever process that will import the
CONVERT_STATSsubworkflow will be able to access and use it's outputs.- Remember inside the
main.nffile for theSAMTOOLS_DEPTHmodule there was this line of code:
- Remember inside the
output:
tuple val(meta1), path("*.tsv"), emit: tsv- This means that the outputs of this module are emitted into an output channel called
tsv. This can be accessed using theoutchannel operator and inserted into the bottom code block.
emit:
bam = ch_bam // channel: [ val(meta), /path/to/bam ] (optional)
bai = ch_bai // channel: [ val(meta), /path/to/bai ] (optional)
cram = ch_cram // channel: [ val(meta), /path/to/cram ] (optional)
crai = ch_crai // channel: [ val(meta), /path/to/crai ] (optional)
depth = SAMTOOLS_DEPTH.out.tsv // channel: [ val(meta), /path/to/tsv ]
stats = SAMTOOLS_STATS.out.stats // channel: [ val(meta), /path/to/stats ]
flagstat = SAMTOOLS_FLAGSTAT.out.flagstat // channel: [ val(meta), /path/to/flagstat ]
idxstats = SAMTOOLS_IDXSTATS.out.idxstats // channel: [ val(meta), /path/to/idxstats ]
versions = ch_versions // channel: [ versions.yml ]
}- Now, with most
nf-corepipelines, these output channels will usually be caught by some almagomating program (aka summarizing program), such as MultiQC. Most modules can be added using the general thought process as outlined above, because they typically are called somewhere deep inside the Nextflow pipeline and then their outputs are propogated upwards to be caught by a summarizing program like MultiQC. However, MultiQC is different because it is a program that sits at the top level of the pipeline and is only called after all of the outputs are propogated upwards and combined to be fed to this program, typically at the end of the pipeline.
The final code for the CONVERT_STATS subworkflow updated with the SAMTOOLS_DEPTH module is as follows:
//
// Convert BAM to CRAM, create index and calculate statistics
//
include { CRUMBLE } from '../../modules/nf-core/crumble/main'
include { SAMTOOLS_VIEW as SAMTOOLS_CRAM } from '../../modules/nf-core/samtools/view/main'
include { SAMTOOLS_VIEW as SAMTOOLS_REINDEX } from '../../modules/nf-core/samtools/view/main'
include { SAMTOOLS_STATS } from '../../modules/nf-core/samtools/stats/main'
include { SAMTOOLS_FLAGSTAT } from '../../modules/nf-core/samtools/flagstat/main'
include { SAMTOOLS_IDXSTATS } from '../../modules/nf-core/samtools/idxstats/main'
include { SAMTOOLS_DEPTH } from '../../modules/nf-core/samtools/depth/main'
workflow CONVERT_STATS {
take:
bam // channel: [ val(meta), /path/to/bam, /path/to/bai ]
fasta // channel: [ val(meta), /path/to/fasta ]
main:
ch_versions = Channel.empty()
// Split outfmt parameter into a list
def outfmt_options = params.outfmt.split(',').collect { it.trim() }
// (Optionally) Compress the quality scores of Illumina and PacBio CCS alignments
if ( params.compression == "crumble" ) {
bam
| branch {
meta, bam ->
run_crumble: meta.datatype == "hic" || meta.datatype == "illumina" || meta.datatype == "pacbio"
no_crumble: true
}
| set { ch_bams }
CRUMBLE ( ch_bams.run_crumble, [], [] )
ch_versions = ch_versions.mix( CRUMBLE.out.versions )
// Convert BAM to CRAM
CRUMBLE.out.bam
| mix( ch_bams.no_crumble )
| map { meta, bam -> [meta, bam, []] }
| set { ch_bams_for_conversion }
} else {
bam
| map { meta, bam -> [meta, bam, []] }
| set { ch_bams_for_conversion }
}
// (Optionally) convert to CRAM if it's specified in outfmt
ch_cram = Channel.empty()
ch_crai = Channel.empty()
if ("cram" in outfmt_options) {
SAMTOOLS_CRAM ( ch_bams_for_conversion, fasta, [] )
ch_versions = ch_versions.mix( SAMTOOLS_CRAM.out.versions.first() )
// Combine CRAM and CRAI into one channel
ch_cram = SAMTOOLS_CRAM.out.cram
ch_crai = SAMTOOLS_CRAM.out.crai
}
// Re-generate BAM index if BAM is in outfmt
def ch_data_for_stats
if ("cram" in outfmt_options) {
ch_data_for_stats = ch_cram.join( ch_crai )
} else {
ch_data_for_stats = ch_bams_for_conversion
}
ch_bam = Channel.empty()
ch_bai = Channel.empty()
if ("bam" in outfmt_options) {
// Re-generate BAM index
SAMTOOLS_REINDEX ( ch_bams_for_conversion, fasta, [] )
ch_versions = ch_versions.mix( SAMTOOLS_REINDEX.out.versions.first() )
// Set the BAM and BAI channels for emission
ch_bam = SAMTOOLS_REINDEX.out.bam
ch_bai = SAMTOOLS_REINDEX.out.bai
// If using BAM for stats, use the reindexed BAM
if ( !("cram" in outfmt_options) ) {
ch_data_for_stats = ch_bam.join ( ch_bai )
}
}
// Calculate Depth using Samtools Depth
ch_data_for_samtools_depth = ch_data_for_stats.map { meta, input, index -> [meta, input] }
ch_intervals = ch_data_for_stats.map { meta, input, index -> [meta, []] }
SAMTOOLS_DEPTH(ch_data_for_samtools_depth, ch_intervals)
ch_versions = ch_versions.mix(SAMTOOLS_DEPTH.out.versions.first())
// Calculate statistics
SAMTOOLS_STATS ( ch_data_for_stats, fasta )
ch_versions = ch_versions.mix( SAMTOOLS_STATS.out.versions.first() )
// Calculate statistics based on flag values
SAMTOOLS_FLAGSTAT ( ch_data_for_stats )
ch_versions = ch_versions.mix( SAMTOOLS_FLAGSTAT.out.versions.first() )
// Calculate index statistics
SAMTOOLS_IDXSTATS ( ch_data_for_stats )
ch_versions = ch_versions.mix( SAMTOOLS_IDXSTATS.out.versions.first() )
emit:
bam = ch_bam // channel: [ val(meta), /path/to/bam ] (optional)
bai = ch_bai // channel: [ val(meta), /path/to/bai ] (optional)
cram = ch_cram // channel: [ val(meta), /path/to/cram ] (optional)
crai = ch_crai // channel: [ val(meta), /path/to/crai ] (optional)
depth = SAMTOOLS_DEPTH.out.tsv // channel: [ val(meta), /path/to/tsv ]
stats = SAMTOOLS_STATS.out.stats // channel: [ val(meta), /path/to/stats ]
flagstat = SAMTOOLS_FLAGSTAT.out.flagstat // channel: [ val(meta), /path/to/flagstat ]
idxstats = SAMTOOLS_IDXSTATS.out.idxstats // channel: [ val(meta), /path/to/idxstats ]
versions = ch_versions // channel: [ versions.yml ]
}After you finish updating the section with your code, make sure you continue to do all the rest of the stuff outlined in the inserting a module guidelines.
Nf-core modules (and subworkflows) are a community driven effort to standardize many bioinformatics and data analysis tools, but sometimes there are errors or irregularities that are published in the module or subworkflow and this can break your code in unexpected ways. This is the case (as of right now) with the SAMTOOLS_DEPTH module.
- The
SAMTOOLS_DEPTHmodule takes in 2 inputs, and it has it's input block structured as follows:
input:
tuple val(meta1), path(bam)
tuple val(meta2), path(intervals)Typically, the name for the variable that contains the metamap should always be meta. If there are more then one inputs, the other input variable should be named something like meta1 or meta2, but meta should always be the name of the first one.
In this case, the metamap had 2 inputs, but the first one was the non-standard meta1. This is an issue because often times this variable name is referenced somewhere, usually in some configuration file. This was the case in this pipeline. Inside the original ./readmapping/conf/modules.config file (the file that describes the configurations for each of the modules used in your pipeline), there is a code snipped that looks like this:
withName: '.*:CONVERT_STATS:SAMTOOLS_.*' {
publishDir = [
path: { "${params.outdir}/read_mapping/${meta.datatype}" },
mode: params.publish_dir_mode,
saveAs: { filename -> filename.equals('versions.yml') ? null : filename }
]
}This block of code can be generally read as "For any modules inside the CONVERT_STATS subworkflow that starts with SAMTOOLS_, make sure to save the output files those module programs generated into the read_mapping/WHATEVER-THE-DATATYPE-OF-YOUR-SAMPLE-WAS directory." This sample datatype is extracted from the metadata in your list. In order for this metadata to be extracted properly, it has to be saved under the meta variable, as this is what's explicitly written in the configuration file. In the case of the SAMTOOLS_DEPTH module that we were working with, since it's metadata is not saved in a variable called meta, but rather meta1, this means that none of the outputs from the samtools depth program will be saved to your outputs.
To fix this, there are a few approaches. Two of them are:
- Change the variable name from
meta1tometadirectly in the./modules/nf-core/samtools/depth/main.nffile.
Note
When editing nf-core modules, it's recommended to use the patch tool. Just editing can be dangerous because if nextflow pulls the module from the nf-core/modules remote repository it can overwrite your local changes.
- Add an additional configuration to accomodate the non-standard irregularities in the module. This is a safer (but more hack-y) option because it doesn't run the risk of being overwritten but code in a remote repository.
withName: '.*:CONVERT_STATS:SAMTOOLS_.*' {
publishDir = [
path: { "${params.outdir}/read_mapping/${meta.datatype}" },
mode: params.publish_dir_mode,
saveAs: { filename -> filename.equals('versions.yml') ? null : filename }
]
}
// Needs separate configuration since nf-core/samtools/depth uses meta1 instead of meta as it's metamap name
withName: '.*:CONVERT_STATS:SAMTOOLS_DEPTH' {
publishDir = [
path: { "${params.outdir}/read_mapping/${meta1.datatype}" },
mode: params.publish_dir_mode,
saveAs: { filename -> filename.equals('versions.yml') ? null : filename }
]
}With this fix, all your program outputs should now be published in the pipeline outputs folder.
This is just one example of fixing an irregularity. Every irregularity will be different but this is just a framework for thinking about where to look and how to solve each problem. With time and practice you get more familiar and efficient with the process and will start writing and editing pipelines with ease.
Here I describe how I added the nf-coremultiqc module to my pipeline.
- As with before, the first thing I did was install the module of interest into my pipeline. This was done using the
nf-core modules installcommand
nf-core modules install multiqc- The next step was to investigate where the
MULTIQCmodule should go.MULTIQCshould always go at the top level of the code and collect all the desired software outputs at the end of the pipeline, but figuring out where this top-level/end of the pipeline is can be trickier than it seems. In nextflow and nf-core, themain.nffile in the top level of the code is always the entrypoint for the pipeline to start. However often times this file doesn't contain anything really meaniningful from a dataprocessing perspective and it might make a call to the main "engine" of the pipeline very early on. Often this is something that was imported from the./workflowsfolder. In this pipeline used there, the top-levelmain.nffile contains this:
#!/usr/bin/env nextflow
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
sanger-tol/readmapping
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
----------------------------------------------------------------------------------------
*/
nextflow.enable.dsl = 2
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
IMPORT FUNCTIONS / MODULES / SUBWORKFLOWS / WORKFLOWS
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
*/
include { READMAPPING } from './workflows/readmapping'
include { PIPELINE_INITIALISATION } from './subworkflows/local/utils_nfcore_readmapping_pipeline'
include { PIPELINE_COMPLETION } from './subworkflows/local/utils_nfcore_readmapping_pipeline'
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
GENOME PARAMETER VALUES
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
*/
workflow SANGERTOL_READMAPPING {
take:
samplesheet
fasta
header
main:
READMAPPING (
samplesheet,
fasta,
header
)
}
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
RUN MAIN WORKFLOW
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
*/
workflow {
main:
//
// SUBWORKFLOW: Run initialisation tasks
//
PIPELINE_INITIALISATION (
params.version,
params.help,
params.validate_params,
params.monochrome_logs,
args,
params.outdir,
params.input
)
//
// WORKFLOW: Run main workflow
//
SANGERTOL_READMAPPING (
PIPELINE_INITIALISATION.out.samplesheet,
PIPELINE_INITIALISATION.out.fasta,
PIPELINE_INITIALISATION.out.header
)
//
// SUBWORKFLOW: Run completion tasks
//
PIPELINE_COMPLETION (
params.email,
params.email_on_fail,
params.plaintext_email,
params.outdir,
params.monochrome_logs,
params.hook_url,
[]
)
}
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
THE END
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
*/-
The
workflowof this file basically makes 3 calls to other smaller pipeline processes:PIPELINE_INITIALISATION,SANGERTOL_READMAPPING, andPIPELINE_COMPLETION. It might be tempting to think that since we need to put theMULTIQCmodule at the end of the pipeline, we should put it in thePIPELINE_COMPLETIONsubworflow, but if you look carefully you'll notice that this workflow has very little to do with handling any biological data and more so with generating logs and emails and things of that nature. All the bulk of the biological data is flowing through theSANGERTOL_READMAPPINGpipeline, which is imported from./workflows/readmapping. -
If we take a look at the
./workflows/readmapping.nffile, we can see the following code:
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
IMPORT LOCAL MODULES/SUBWORKFLOWS
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
*/
//
// MODULE: Local modules
//
include { SAMTOOLS_REHEADER } from '../modules/local/samtools_replaceheader'
//
// SUBWORKFLOW: Consisting of a mix of local and nf-core/modules
//
include { INPUT_CHECK } from '../subworkflows/local/input_check'
include { PREPARE_GENOME } from '../subworkflows/local/prepare_genome'
include { ALIGN_SHORT as ALIGN_HIC } from '../subworkflows/local/align_short'
include { ALIGN_SHORT as ALIGN_ILLUMINA } from '../subworkflows/local/align_short'
include { ALIGN_PACBIO as ALIGN_HIFI } from '../subworkflows/local/align_pacbio'
include { ALIGN_PACBIO as ALIGN_CLR } from '../subworkflows/local/align_pacbio'
include { ALIGN_ONT } from '../subworkflows/local/align_ont'
include { CONVERT_STATS } from '../subworkflows/local/convert_stats'
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
IMPORT NF-CORE MODULES/SUBWORKFLOWS
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
*/
//
// MODULE: Installed directly from nf-core/modules
//
include { UNTAR } from '../modules/nf-core/untar/main'
include { CUSTOM_DUMPSOFTWAREVERSIONS } from '../modules/nf-core/custom/dumpsoftwareversions/main'
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
RUN MAIN WORKFLOW
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
*/
workflow READMAPPING {
take:
ch_samplesheet
ch_fasta
ch_header
main:
// Initialize an empty versions channel
ch_versions = Channel.empty()
ch_multiqc_files = Channel.empty()
//
// SUBWORKFLOW: Read in samplesheet, validate and stage input files
//
INPUT_CHECK ( ch_samplesheet ).reads
| branch {
meta, reads ->
hic : meta.datatype == "hic"
illumina : meta.datatype == "illumina"
pacbio : meta.datatype == "pacbio"
clr : meta.datatype == "pacbio_clr"
ont : meta.datatype == "ont"
}
| set { ch_reads }
ch_versions = ch_versions.mix ( INPUT_CHECK.out.versions )
ch_fasta
| map { [ [ id: it.baseName ], it ] }
| set { ch_genome }
PREPARE_GENOME ( ch_genome )
ch_versions = ch_versions.mix ( PREPARE_GENOME.out.versions )
//
// Create channel for vector DB
//
// ***PacBio condition does not work - needs fixing***
if ( ch_reads.pacbio || ch_reads.clr ) {
if ( params.vector_db.endsWith( '.tar.gz' ) ) {
UNTAR ( [ [:], params.vector_db ] ).untar
| set { ch_vector_db }
ch_versions = ch_versions.mix ( UNTAR.out.versions )
} else {
Channel.fromPath ( params.vector_db )
| set { ch_vector_db }
}
}
//
// SUBWORKFLOW: Align raw reads to genome
//
ALIGN_HIC ( PREPARE_GENOME.out.fasta, PREPARE_GENOME.out.bwaidx, ch_reads.hic )
ch_versions = ch_versions.mix ( ALIGN_HIC.out.versions )
ALIGN_ILLUMINA ( PREPARE_GENOME.out.fasta, PREPARE_GENOME.out.bwaidx, ch_reads.illumina )
ch_versions = ch_versions.mix ( ALIGN_ILLUMINA.out.versions )
ALIGN_HIFI ( PREPARE_GENOME.out.fasta, ch_reads.pacbio, ch_vector_db )
ch_versions = ch_versions.mix ( ALIGN_HIFI.out.versions )
ALIGN_CLR ( PREPARE_GENOME.out.fasta, ch_reads.clr, ch_vector_db )
ch_versions = ch_versions.mix ( ALIGN_CLR.out.versions )
ALIGN_ONT ( PREPARE_GENOME.out.fasta, ch_reads.ont )
ch_versions = ch_versions.mix ( ALIGN_ONT.out.versions )
// gather alignments
ch_aligned_bams = Channel.empty()
| mix( ALIGN_HIC.out.bam )
| mix( ALIGN_ILLUMINA.out.bam )
| mix( ALIGN_HIFI.out.bam )
| mix( ALIGN_CLR.out.bam )
| mix( ALIGN_ONT.out.bam )
// Optionally insert params.header information to bams
ch_reheadered_bams = Channel.empty()
if ( params.header ) {
SAMTOOLS_REHEADER( ch_aligned_bams, ch_header.first() )
ch_reheadered_bams = SAMTOOLS_REHEADER.out.bam
ch_versions = ch_versions.mix ( SAMTOOLS_REHEADER.out.versions )
} else {
ch_reheadered_bams = ch_aligned_bams
}
// convert to cram and gather stats
CONVERT_STATS ( ch_reheadered_bams, PREPARE_GENOME.out.fasta )
ch_versions = ch_versions.mix ( CONVERT_STATS.out.versions )
//
// MODULE: Combine different versions.yml
//
CUSTOM_DUMPSOFTWAREVERSIONS (
ch_versions
| unique { it.text }
| collectFile ( name: 'collated_versions.yml' )
)
}
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
THE END
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
*/- Here at the end you can see a call to
CUSTOM_DUMPSOFTWAREVERSIONS. Emitting the versions of all the softwares used in this pipeline usally indicates that this is the end of the dataprocessing, because all the programs have run and their software versions have been collected. This indicates this we can probably add our call to themultiqcprogram somewhere here. Now that we've determined the endpoint of the data processing in the pipeline, we can begin working on adding theMULTIQCmodule here.
- The next step is to inspect the
MULTIQCmodule that we installed (inside the./modules/nf-core/multiqc/main.nffile) to understand what inputs it takes and outputs it emits. The module looks like this:
process MULTIQC {
label 'process_single'
conda "${moduleDir}/environment.yml"
container "${ workflow.containerEngine == 'singularity' && !task.ext.singularity_pull_docker_container ?
'https://depot.galaxyproject.org/singularity/multiqc:1.25.1--pyhdfd78af_0' :
'biocontainers/multiqc:1.25.1--pyhdfd78af_0' }"
input:
path multiqc_files, stageAs: "?/*"
path(multiqc_config)
path(extra_multiqc_config)
path(multiqc_logo)
path(replace_names)
path(sample_names)
output:
path "*multiqc_report.html", emit: report
path "*_data" , emit: data
path "*_plots" , optional:true, emit: plots
path "versions.yml" , emit: versions
when:
task.ext.when == null || task.ext.when
script:
def args = task.ext.args ?: ''
def prefix = task.ext.prefix ? "--filename ${task.ext.prefix}.html" : ''
def config = multiqc_config ? "--config $multiqc_config" : ''
def extra_config = extra_multiqc_config ? "--config $extra_multiqc_config" : ''
def logo = multiqc_logo ? "--cl-config 'custom_logo: \"${multiqc_logo}\"'" : ''
def replace = replace_names ? "--replace-names ${replace_names}" : ''
def samples = sample_names ? "--sample-names ${sample_names}" : ''
"""
multiqc \\
--force \\
$args \\
$config \\
$prefix \\
$extra_config \\
$logo \\
$replace \\
$samples \\
.
cat <<-END_VERSIONS > versions.yml
"${task.process}":
multiqc: \$( multiqc --version | sed -e "s/multiqc, version //g" )
END_VERSIONS
"""
stub:
"""
mkdir multiqc_data
mkdir multiqc_plots
touch multiqc_report.html
cat <<-END_VERSIONS > versions.yml
"${task.process}":
multiqc: \$( multiqc --version | sed -e "s/multiqc, version //g" )
END_VERSIONS
"""
}- You can see that the
MULTIQCmodule takes in alot of inputs. If you inspect them closely however, you'll notice that all of them are optional, with the exception of the first onepath multiqc_files, stageAs: "?/*"(Remember that if you see a pattern like thisinput_variable ? "--some_parameter $input_variable" : ''in thescriptblock, that indicates this input was optional). This means that we can focus on preparing the first input, and then feed the module dummy channels to satisfy the other input requirments.
- Now knowing what we know, it's time to insert the
MULTIQCmodule at the end of the workflow. The first thing to do is to import the module into the./workflows/readmapping.nfworkflow.
//
// MODULE: Installed directly from nf-core/modules
//
include { UNTAR } from '../modules/nf-core/untar/main'
include { CUSTOM_DUMPSOFTWAREVERSIONS } from '../modules/nf-core/custom/dumpsoftwareversions/main'
include { MULTIQC } from '../modules/nf-core/multiqc'- Next it's time to create the mandatory input channel for the
MULTIQCmodule. We can start of as setting this to an empty channel for now:
main:
// Initialize an empty versions channel
ch_versions = Channel.empty()
ch_multiqc_files = Channel.empty()-
Now it's important to fill this channel with all the data we want in our final output. The issue is that the data we want is being emitted in the
CONVERT_STATSsubworkflow. If we want that data in thisch_multiqc_fileschannel, we need a way to propograte it from theCONVERT_STATSsubworkflow to theREADMAPPINGworkflow. This means that we will have to go back into theCONVERT_STATSsubworkflow code and modify it to accomodate capturing data to be used in the MultiQC report. -
So back in the
./readmapping/subworkflows/local/convert_stats.nfsubworkflow, initialize another channel for multiqc files (ch_multiqc_files).
workflow CONVERT_STATS {
take:
bam // channel: [ val(meta), /path/to/bam, /path/to/bai ]
fasta // channel: [ val(meta), /path/to/fasta ]
main:
ch_versions = Channel.empty()
ch_multiqc_files = Channel.empty()- Then for every process that generates an output, add a line that captures that output inside the multiqc channel. The Nextflow channel operator
mixis really useful for this purpose.
// Calculate Depth using Samtools Depth
ch_data_for_samtools_depth = ch_data_for_stats.map { meta, input, index -> [meta, input] }
ch_intervals = ch_data_for_stats.map { meta, input, index -> [meta, []] }
SAMTOOLS_DEPTH(ch_data_for_samtools_depth, ch_intervals)
ch_versions = ch_versions.mix(SAMTOOLS_DEPTH.out.versions.first())
ch_multiqc_files = SAMTOOLS_DEPTH.out.tsv.mix(ch_multiqc_files)
// Calculate statistics
SAMTOOLS_STATS ( ch_data_for_stats, fasta )
ch_versions = ch_versions.mix( SAMTOOLS_STATS.out.versions.first() )
ch_multiqc_files = SAMTOOLS_STATS.out.stats.mix(ch_multiqc_files)
// Calculate statistics based on flag values
SAMTOOLS_FLAGSTAT ( ch_data_for_stats )
ch_versions = ch_versions.mix( SAMTOOLS_FLAGSTAT.out.versions.first() )
ch_multiqc_files = SAMTOOLS_FLAGSTAT.out.flagstat.mix(ch_multiqc_files)
// Calculate index statistics
SAMTOOLS_IDXSTATS ( ch_data_for_stats )
ch_versions = ch_versions.mix( SAMTOOLS_IDXSTATS.out.versions.first() )
ch_multiqc_files = SAMTOOLS_IDXSTATS.out.idxstats.mix(ch_multiqc_files)Note
If you have a module or a process that produces multiple outputs and you are trying to capture all of them, you can chain together many mix operations. For example, when I was working on editing this pipeline myself, in one version I added the MOSDEPTH module to this subworkflow. This is how I did it:
// Calculate sequencing depth using mosdepth
ch_data_for_mosdepth = ch_data_for_stats.map { meta, input, index -> [meta, input, index, []] }
MOSDEPTH(ch_data_for_mosdepth, fasta)
ch_versions = ch_versions.mix(MOSDEPTH.out.versions.first())
ch_multiqc_files = MOSDEPTH.out.global_txt
.mix(MOSDEPTH.out.summary_txt)
.mix(MOSDEPTH.out.regions_txt)
.mix(MOSDEPTH.out.per_base_d4)
.mix(MOSDEPTH.out.per_base_bed)
.mix(MOSDEPTH.out.per_base_csi)
.mix(MOSDEPTH.out.regions_bed)
.mix(MOSDEPTH.out.regions_csi)
.mix(MOSDEPTH.out.quantized_bed)
.mix(MOSDEPTH.out.quantized_csi)
.mix(MOSDEPTH.out.thresholds_bed)
.mix(MOSDEPTH.out.thresholds_csi)
.mix(ch_multiqc_files)- Once this is complete, the last thing to do is to make sure you propogate up this multiqc channel (in other words, emit the multiqc channel so that it is able to be used in a workflow that imports this subworkflow).
- Because this is a special channel, you may have to modify it in a special way to get the output correct. A useful operator for this is the
transponse()function. Note that this function is not the Nextflowtransposechannel operator, but rather thetransposemethod of Groovy collections. Remember that Nextflow, Groovy, and Java are all interconnected and sometimes it might be useful to use a function from one of the lower level languages to get your desired effect, as is often the case with thetranspose()method. This one is just extra confusing becasue it shares a name with a Nextflow operator. Often times you can tell the difference because the Groovy method will just be invoke with empty parentheses()(e.g.transpose()).
- Because this is a special channel, you may have to modify it in a special way to get the output correct. A useful operator for this is the
emit:
bam = ch_bam // channel: [ val(meta), /path/to/bam ] (optional)
bai = ch_bai // channel: [ val(meta), /path/to/bai ] (optional)
cram = ch_cram // channel: [ val(meta), /path/to/cram ] (optional)
crai = ch_crai // channel: [ val(meta), /path/to/crai ] (optional)
depth = SAMTOOLS_DEPTH.out.tsv // channel: [ val(meta), /path/to/tsv ]
stats = SAMTOOLS_STATS.out.stats // channel: [ val(meta), /path/to/stats ]
flagstat = SAMTOOLS_FLAGSTAT.out.flagstat // channel: [ val(meta), /path/to/flagstat ]
idxstats = SAMTOOLS_IDXSTATS.out.idxstats // channel: [ val(meta), /path/to/idxstats ]
versions = ch_versions // channel: [ versions.yml ]
multiqc_files = ch_multiqc_files.transpose().map { it[1] }Note
This has to be done in every subworkflow in which you want data to appear in the MultiQC report. In this specific example, we are only concerned about the data that is outputted in the CONVERT_STATS subworkflow.
-
Now that the output data is being propogated upwards from our desired subworkflows, we can return back to our workflow and continue implementing the
MULTIQCmodule. -
So back in the
./readmapping/workflows/readmapping.nffile, collect multiqc channel data emitted by theCONVERT_STATSsubworkflow:
ch_versions = ch_versions.mix ( CONVERT_STATS.out.versions )
ch_multiqc_files = ch_multiqc_files.mix(CONVERT_STATS.out.multiqc_files)- Now you're ready to feed this data into the
MULTIQCmodule. Before you do so, make sure the MultiQC channel is formatted correctly. Thecollectoperator puts everything into a single list entry which can then be fed intoMULTIQC:
// Also can load some MultiQC configuration files before calling the program
ch_multiqc_custom_config = params.multiqc_config ? Channel.fromPath(params.multiqc_config) : Channel.empty() // CAN ALSO DO SOMETHING LIKE Channel.fromPath("$projectDir/workflows/assets/multiqc/multiqc_config.yml", checkIfExists: true) IF YOU HAVE ANY CONFIGURATION FILES HERE
ch_multiqc_logo = params.multiqc_logo ? Channel.fromPath(params.multiqc_logo) : Channel.empty()
// Call MultiQC program
// Remember the inputs:
// input:
// path multiqc_files, stageAs: "?/*"
// path(multiqc_config)
// path(extra_multiqc_config)
// path(multiqc_logo)
// path(replace_names)
// path(sample_names)
MULTIQC (
ch_multiqc_files.collect(),
[],
ch_multiqc_custom_config.toList(),
ch_multiqc_logo.toList(),
[],
[] // empty lists are dummy channels
)- Now we're almost done. The
MULTIQCprogram has been executed and all the important data has been summarized, but remember how I mentionned that there were three workflows that were run in the Nextflow entrypoint (the top-levelmain.nffile):PIPELINE_INITIALISATION,SANGERTOL_READMAPPING, andPIPELINE_COMPLETION? Even thoughSANGERTOL_READMAPPINGis where all the important data processing and program executions takes place, it's still technically not the end of the whole pipeline. Nextflow requires you to explicitly declare if you want a program's outputs pubished. This includes theMULTIQCprogram. There is a chance there is some important code that publishes theMULTIQCresults to your outputs folder in the last stage of the pipeline, thePIPELINE_COMPLETIONsubworkflow, and so it's a really good idea to check whether or not there is anything in that subworkflow's code that indicates we should propogate theMULTIQCresults there.- Inside the
./subworkflows/local/utils_nfcore_readmapping_pipeline/main.nffile, you can find the following code snippet:
- Inside the
workflow PIPELINE_COMPLETION {
take:
email // string: email address
email_on_fail // string: email address sent on pipeline failure
plaintext_email // boolean: Send plain-text email instead of HTML
outdir // path: Path to output directory where results will be published
monochrome_logs // boolean: Disable ANSI colour codes in log output
hook_url // string: hook URL for notifications
multiqc_report // string: Path to MultiQC report- Here you can see that one of the things the
PIPELINE_COMPLETIONsubworkflow takes as input a channel calledmultiqc_report, which contains the path to the MultiQC report. This suggests that in ourSANGERTOL_READMAPPINGworkflow, we should emit our multiqc output. So back in our./workflows/readmapping.nffile, after calling theMULTIQCprogram, we should add the following code:
// Save the path to the MultiQC report in a channel
// Remember from the output block of the rom the MULTIQC module that the path to the MultiQC report is emitted as `report`
//
// output:
// path "*multiqc_report.html", emit: report
ch_multiqc_report = MULTIQC.out.report
// Emit that channel so that it can be captured by the PIPELINE_COMPLETION subworkflow
// Make sure to name the emission the same thing that the PIPELINE_COMPLETION takes as input: multiqc_report
emit:
multiqc_report = ch_multiqc_report // channel: /path/to/multiqc_report.html- And that's it. Now you can test your pipeline to see if it works. From here you can go and read the rest of the instructions outlined in the inserting a module guidelines to see what else you need to do to follow the best practices. MultiQC also comes with a lot of configutation options. You can read the documention and look at other nf-core pipelines to see how they implemented different configuration options. It is a huge module and too much to go into right here but with this knowledge you should be ready to go on and pursue more knowledge yourself.
-
To summarize, this is what the final files look like after successfully adding the
MULTIQCmodule. -
./subworkflows/local/convert_stats.nf
//
// Convert BAM to CRAM, create index and calculate statistics
//
include { CRUMBLE } from '../../modules/nf-core/crumble/main'
include { SAMTOOLS_VIEW as SAMTOOLS_CRAM } from '../../modules/nf-core/samtools/view/main'
include { SAMTOOLS_VIEW as SAMTOOLS_REINDEX } from '../../modules/nf-core/samtools/view/main'
include { SAMTOOLS_STATS } from '../../modules/nf-core/samtools/stats/main'
include { SAMTOOLS_FLAGSTAT } from '../../modules/nf-core/samtools/flagstat/main'
include { SAMTOOLS_IDXSTATS } from '../../modules/nf-core/samtools/idxstats/main'
include { SAMTOOLS_DEPTH } from '../../modules/nf-core/samtools/depth/main'
workflow CONVERT_STATS {
take:
bam // channel: [ val(meta), /path/to/bam, /path/to/bai ]
fasta // channel: [ val(meta), /path/to/fasta ]
main:
ch_versions = Channel.empty()
ch_multiqc_files = Channel.empty()
// Split outfmt parameter into a list
def outfmt_options = params.outfmt.split(',').collect { it.trim() }
// (Optionally) Compress the quality scores of Illumina and PacBio CCS alignments
if (params.compression == "crumble") {
bam
| branch { meta, bam ->
run_crumble: meta.datatype == "hic" || meta.datatype == "illumina" || meta.datatype == "pacbio"
no_crumble: true
}
| set { ch_bams }
CRUMBLE(ch_bams.run_crumble, [], [])
ch_versions = ch_versions.mix(CRUMBLE.out.versions)
// Convert BAM to CRAM
CRUMBLE.out.bam
| mix(ch_bams.no_crumble)
| map { meta, bam -> [meta, bam, []] }
| set { ch_bams_for_conversion }
}
else {
bam
| map { meta, bam -> [meta, bam, []] }
| set { ch_bams_for_conversion }
}
// (Optionally) convert to CRAM if it's specified in outfmt
ch_cram = Channel.empty()
ch_crai = Channel.empty()
if ("cram" in outfmt_options) {
SAMTOOLS_CRAM(ch_bams_for_conversion, fasta, [])
ch_versions = ch_versions.mix(SAMTOOLS_CRAM.out.versions.first())
// Combine CRAM and CRAI into one channel
ch_cram = SAMTOOLS_CRAM.out.cram
ch_crai = SAMTOOLS_CRAM.out.crai
}
// Re-generate BAM index if BAM is in outfmt
def ch_data_for_stats =
if ("cram" in outfmt_options) {
ch_data_for_stats = ch_cram.join(ch_crai)
}
else {
ch_data_for_stats = ch_bams_for_conversion
}
ch_bam = Channel.empty()
ch_bai = Channel.empty()
if ("bam" in outfmt_options) {
// Re-generate BAM index
SAMTOOLS_REINDEX(ch_bams_for_conversion, fasta, [])
ch_versions = ch_versions.mix(SAMTOOLS_REINDEX.out.versions.first())
// Set the BAM and BAI channels for emission
ch_bam = SAMTOOLS_REINDEX.out.bam
ch_bai = SAMTOOLS_REINDEX.out.bai
// If using BAM for stats, use the reindexed BAM
if (!("cram" in outfmt_options)) {
ch_data_for_stats = ch_bam.join(ch_bai)
}
}
// Index fasta
ch_fai = Channel.empty()
SAMTOOLS_FAIDX(fasta, [[:], []])
ch_fai = SAMTOOLS_FAIDX.out.fai
ch_versions = ch_versions.mix(SAMTOOLS_FAIDX.out.versions.first())
// Calculate Depth using Samtools Depth
ch_data_for_samtools_depth = ch_data_for_stats.map { meta, input, index -> [meta, input] }
ch_intervals = ch_data_for_stats.map { meta, input, index -> [meta, []] }
SAMTOOLS_DEPTH(ch_data_for_samtools_depth, ch_intervals)
ch_versions = ch_versions.mix(SAMTOOLS_DEPTH.out.versions.first())
ch_multiqc_files = SAMTOOLS_DEPTH.out.tsv.mix(ch_multiqc_files)
// Calculate statistics
SAMTOOLS_STATS(ch_data_for_stats, fasta)
ch_versions = ch_versions.mix(SAMTOOLS_STATS.out.versions.first())
ch_multiqc_files = SAMTOOLS_STATS.out.stats.mix(ch_multiqc_files)
// Calculate statistics based on flag values
SAMTOOLS_FLAGSTAT(ch_data_for_stats)
ch_versions = ch_versions.mix(SAMTOOLS_FLAGSTAT.out.versions.first())
ch_multiqc_files = SAMTOOLS_FLAGSTAT.out.flagstat.mix(ch_multiqc_files)
// Calculate index statistics
SAMTOOLS_IDXSTATS(ch_data_for_stats)
ch_versions = ch_versions.mix(SAMTOOLS_IDXSTATS.out.versions.first())
ch_multiqc_files = SAMTOOLS_IDXSTATS.out.idxstats.mix(ch_multiqc_files)
emit:
bam = ch_bam // channel: [ val(meta), /path/to/bam ] (optional)
bai = ch_bai // channel: [ val(meta), /path/to/bai ] (optional)
cram = ch_cram // channel: [ val(meta), /path/to/cram ] (optional)
crai = ch_crai // channel: [ val(meta), /path/to/crai ] (optional)
depth = SAMTOOLS_DEPTH.out.tsv // channel: [ val(meta), /path/to/tsv ]
stats = SAMTOOLS_STATS.out.stats // channel: [ val(meta), /path/to/stats ]
flagstat = SAMTOOLS_FLAGSTAT.out.flagstat // channel: [ val(meta), /path/to/flagstat ]
idxstats = SAMTOOLS_IDXSTATS.out.idxstats // channel: [ val(meta), /path/to/idxstats ]
versions = ch_versions // channel: [ versions.yml ]
multiqc_files = ch_multiqc_files.transpose().map { it[1] }
}
./subworkflows/local/utils_nfcore_readmapping_pipeline/main.nf
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
IMPORT LOCAL MODULES/SUBWORKFLOWS
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
*/
//
// MODULE: Local modules
//
include { SAMTOOLS_REHEADER } from '../modules/local/samtools_replaceheader'
//
// SUBWORKFLOW: Consisting of a mix of local and nf-core/modules
//
include { INPUT_CHECK } from '../subworkflows/local/input_check'
include { PREPARE_GENOME } from '../subworkflows/local/prepare_genome'
include { ALIGN_SHORT as ALIGN_HIC } from '../subworkflows/local/align_short'
include { ALIGN_SHORT as ALIGN_ILLUMINA } from '../subworkflows/local/align_short'
include { ALIGN_PACBIO as ALIGN_HIFI } from '../subworkflows/local/align_pacbio'
include { ALIGN_PACBIO as ALIGN_CLR } from '../subworkflows/local/align_pacbio'
include { ALIGN_ONT } from '../subworkflows/local/align_ont'
include { CONVERT_STATS } from '../subworkflows/local/convert_stats'
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
IMPORT NF-CORE MODULES/SUBWORKFLOWS
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
*/
//
// MODULE: Installed directly from nf-core/modules
//
include { UNTAR } from '../modules/nf-core/untar/main'
include { CUSTOM_DUMPSOFTWAREVERSIONS } from '../modules/nf-core/custom/dumpsoftwareversions/main'
include { MULTIQC } from '../modules/nf-core/multiqc'
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
RUN MAIN WORKFLOW
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
*/
// Header files for MultiQC (TODO)
workflow READMAPPING {
take:
ch_samplesheet
ch_fasta
ch_header
main:
// Initialize an empty versions channel
ch_versions = Channel.empty()
ch_multiqc_files = Channel.empty()
//
// SUBWORKFLOW: Read in samplesheet, validate and stage input files
//
INPUT_CHECK ( ch_samplesheet ).reads
| branch {
meta, reads ->
hic : meta.datatype == "hic"
illumina : meta.datatype == "illumina"
pacbio : meta.datatype == "pacbio"
clr : meta.datatype == "pacbio_clr"
ont : meta.datatype == "ont"
}
| set { ch_reads }
ch_versions = ch_versions.mix ( INPUT_CHECK.out.versions )
ch_fasta
| map { [ [ id: it.baseName ], it ] }
| set { ch_genome }
PREPARE_GENOME ( ch_genome )
ch_versions = ch_versions.mix ( PREPARE_GENOME.out.versions )
//
// Create channel for vector DB
//
// ***PacBio condition does not work - needs fixing***
if ( ch_reads.pacbio || ch_reads.clr ) {
if ( params.vector_db.endsWith( '.tar.gz' ) ) {
UNTAR ( [ [:], params.vector_db ] ).untar
| set { ch_vector_db }
ch_versions = ch_versions.mix ( UNTAR.out.versions )
} else {
Channel.fromPath ( params.vector_db )
| set { ch_vector_db }
}
}
//
// SUBWORKFLOW: Align raw reads to genome
//
ALIGN_HIC ( PREPARE_GENOME.out.fasta, PREPARE_GENOME.out.bwaidx, ch_reads.hic )
ch_versions = ch_versions.mix ( ALIGN_HIC.out.versions )
ALIGN_ILLUMINA ( PREPARE_GENOME.out.fasta, PREPARE_GENOME.out.bwaidx, ch_reads.illumina )
ch_versions = ch_versions.mix ( ALIGN_ILLUMINA.out.versions )
ALIGN_HIFI ( PREPARE_GENOME.out.fasta, ch_reads.pacbio, ch_vector_db )
ch_versions = ch_versions.mix ( ALIGN_HIFI.out.versions )
ALIGN_CLR ( PREPARE_GENOME.out.fasta, ch_reads.clr, ch_vector_db )
ch_versions = ch_versions.mix ( ALIGN_CLR.out.versions )
ALIGN_ONT ( PREPARE_GENOME.out.fasta, ch_reads.ont )
ch_versions = ch_versions.mix ( ALIGN_ONT.out.versions )
// gather alignments
ch_aligned_bams = Channel.empty()
| mix( ALIGN_HIC.out.bam )
| mix( ALIGN_ILLUMINA.out.bam )
| mix( ALIGN_HIFI.out.bam )
| mix( ALIGN_CLR.out.bam )
| mix( ALIGN_ONT.out.bam )
// Optionally insert params.header information to bams
ch_reheadered_bams = Channel.empty()
if ( params.header ) {
SAMTOOLS_REHEADER( ch_aligned_bams, ch_header.first() )
ch_reheadered_bams = SAMTOOLS_REHEADER.out.bam
ch_versions = ch_versions.mix ( SAMTOOLS_REHEADER.out.versions )
} else {
ch_reheadered_bams = ch_aligned_bams
}
// convert to cram and gather stats
CONVERT_STATS ( ch_reheadered_bams, PREPARE_GENOME.out.fasta )
ch_versions = ch_versions.mix ( CONVERT_STATS.out.versions )
ch_multiqc_files = ch_multiqc_files.mix(CONVERT_STATS.out.multiqc_files)
//
// MODULE: Combine different versions.yml
//
CUSTOM_DUMPSOFTWAREVERSIONS (
ch_versions
| unique { it.text }
| collectFile ( name: 'collated_versions.yml' )
)
// Load MultiQC configuration files
ch_multiqc_custom_config = params.multiqc_config ? Channel.fromPath(params.multiqc_config) : Channel.empty() // CAN ALSO DO SOMETHING LIKE Channel.fromPath("$projectDir/workflows/assets/multiqc/multiqc_config.yml", checkIfExists: true) IF YOU HAVE ANY CONFIGURATION FILES HERE
ch_multiqc_logo = params.multiqc_logo ? Channel.fromPath(params.multiqc_logo) : Channel.empty()
// Call MultiQC program
// Remember the inputs:
// input:
// path multiqc_files, stageAs: "?/*"
// path(multiqc_config)
// path(extra_multiqc_config)
// path(multiqc_logo)
// path(replace_names)
// path(sample_names)
MULTIQC (
ch_multiqc_files.collect(),
[], // empty lists are dummy channels
ch_multiqc_custom_config.toList(),
ch_multiqc_logo.toList(),
[],
[]
)
ch_multiqc_report = MULTIQC.out.report
emit:
multiqc_report = ch_multiqc_report // channel: /path/to/multiqc_report.html
}
/*
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
THE END
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
*/Nextflow is a domain specific language (DSL) implemented on top of the Groovy programming language, which in turn is a super-set of the Java programming language. This means that Nextflow can run any Groovy or Java code.
In some cases it may be useful to know a bit of Groovy when building Nextflow pipelines whenever Nextflow functions are insufficient (rare but sometimes may be the case).
Here are some important things to know about Groovy.
To define a variable, simply assign a value to it:
x = 1These variables are global variables.
To define a local variable, use the def keyword:
def x = 1The def should be always used when defining variables local to a function or a closure.
Groovy supports the use of the ternary operator.
def x = condition ? valueIfTrue : valueIfFalseGroovy supports the use of the Elvis operator (?:).
somethingThatMightBeFalsey ?: somethingElse
// ^This is equivalent to
somethingThatMightBeFalsey ? somethingThatMightBeFalsey : somethingElseAnother form of the Elvis operator can be used to quickly assign a conditional value to a variable (?=).
meta.id ?= meta.sample
// ^This is equivalent to
meta.id = meta.id ? meta.id : meta.sampleYou can use the assert keyword to test if a condition is true (similar to an if function).
Here, Groovy will print nothing if it is correct, else it will raise an AssertionError message.
list = [10, 20, 30, 40]
assert list[0] == 10 // Nothing happens
assert list[0] == 20 /* Outputs
ERROR ~ assert list[0] == 20
| |
| 10
[10, 20, 30, 40]
*/String literals can be defined by enclosing them with either single- ('') or double- ("") quotation marks.
- Strings enclosed in single quotes are treated literally, meaning they are not subject to string interpolation.
- This includes escape characters such as
\n,\t,\\, etc. They will be treated literally.
- This includes escape characters such as
- Strings enclosed in double quotes allow string interpolation, meaning variables and expressions within the string are evaluated and replaced with their values.
Example:
x = 'Hello'
y = 'World'
println '$x $y' // Outputs: $x $y
println "$x $y" // Outputs: Hello WorldBelow is another example of how strings can be constructed using string interpolation:
foxtype = 'quick'
foxcolor = ['b', 'r', 'o', 'w', 'n']
println "The $foxtype ${foxcolor.join()} fox"
// Outputs: The quick brown foxNote the different use of $ and ${..} syntax to interpolate value expressions in a string literal.
Groovy has a lot of very helpful syntactic sugar for string manipulation.
You can trim parts of a string by simply subtracting another string.
Example:
demo = "one two three"
assertEquals(demo - "two ", "one three")Regex's can be creating in Groovy using the ~ pattern
demo = "one two three"
assertEquals(demo - ~/t.o ?/, "one three")Note
In Groovy, - is just a shorthand notation for the minus method.
The tokenize() method is a convenient method provided by Groovy for splitting strings into tokens based on a delimiter (kind of like the .split() method in Python). It is really useful for parsing metadata delimited by a certain delimeter.
Example:
def metadata = "sampleA_rep1_tumor"
tokens = metadata.tokenize("_")
println(tokens)
// Outputs: [sampleA, rep1, tumor]Combining this with destructuring syntax you can quickly create a lot of variables for your metadata.
Example:
def metadata = "sampleA_rep1_tumor"
(sample, replicate, type) = metadata.tokenize("_")
println(sample)
println(replicate)
println(type)
/* Outputs:
sampleA
rep1
tumor
*/Note
Be aware of an important difference between the tokenize and split methods in Groovy. The tokenize() method uses each character of a string as a delimiter whereas split() takes the entire string as a delimiter.
Example:
String testString='hello world'
assert ['hel',' world']==testString.split('lo')
assert ['he',' w','r','d']==testString.tokenize('lo')Maps are like lists that have an arbitrary key instead of an integer. Therefore, the syntax is very much aligned.
map = [a: 0, b: 1, c: 2]Maps can be accessed in a conventional square-bracket syntax or as if the key was a property of the map.
map = [a: 0, b: 1, c: 2]
assert map['a'] == 0
assert map.b == 1
assert map.get('c') == 2To add data or to modify a map, the syntax is similar to adding values to a list:
map = [a: 0, b: 1, c: 2]
map['a'] = 'x'
map.b = 'y'
map.put('c', 'z')
assert map == [a: 'x', b: 'y', c: 'z']Map objects implement all methods provided by the java.util.Map interface, plus the extension methods provided by Groovy.
The keySet() method is a Java method (not a Groovy method), but it's still very useful when working with map objects in Groovy and Nextflow. It is part of the Map interface, which is implemented by classes like HashMap, TreeMap, etc. This method returns a Set containing all the keys present in the map.
Often in Nextflow you'll have your metadata being contained in a map. If you want to extract the names of the keys this method turns out to be very convienient.
Example:
(readKeys, metaKeys) = row.keySet().split { it =~ /^fastq/ }It is possible to define a custom function into a script:
def fib(int n) {
return n < 2 ? 1 : fib(n - 1) + fib(n - 2)
}
assert fib(10)==89A function can take multiple arguments separating them with a comma.
The return keyword can be omitted and the function implicitly returns the value of the last evaluated expression. Also, explicit types can be omitted, though not recommended:
def fact(n) {
n > 1 ? n * fact(n - 1) : 1
}
assert fact(5) == 120In Groovy, closures are blocks of code that can take arguments, execute code, and return a value. These blocks of code can be passed as an argument to a function. They are similar to anonymous functions or lambda expressions in other programming languages.
More formally, a closure allows the definition of functions as first-class objects.
square = { it * it }The curly brackets around the expression it * it tells the script interpreter to treat this expression as code. The it identifier is an implicit variable that represents the value that is passed to the function when it is invoked.
Once compiled, the function object is assigned to the variable square as any other variable assignment shown previously.
To invoke the closure execution use the special method call or just use the round parentheses to specify the closure parameter(s):
assert square.call(5) == 25
assert square(9) == 81As is, this may not seem interesting, but you can now pass the square function as an argument to other functions or methods. Some built-in functions take a function like this as an argument. One example is the collect method on lists:
x = [1, 2, 3, 4].collect(square)
println x
// Outputs: [1, 4, 9, 16]Note
This collect method in groovy is different than the collect operator in Nextflow. This groovy collect method does something to each element of the list. In Nextflow there is a map operator that performs the same function.)
By default, closures take a single parameter called it.
To give it a different name use the -> syntax. For example:
square = { num -> num * num }It’s also possible to define closures with multiple, custom-named parameters.
For example, when the method each() is applied to a map it can take a closure with two arguments, to which it passes the key-value pair for each entry in the map object.
Example:
printMap = { a, b -> println "$a with value $b" }
values = ["Yue": "Wu", "Mark": "Williams", "Sudha": "Kumari"]
values.each(printMap) /* Outputs
Yue with value Wu
Mark with value Williams
Sudha with value Kumari
*/A closure has two other important features.
-
It can access and modify variables in the scope where it is defined.
-
A closure can be defined in an anonymous manner, meaning that it is not given a name, and is only defined in the place where it needs to be used.
As an example showing both these features, see the following code fragment:
result = 0
values = ["China": 1, "India": 2, "USA": 3]
values.keySet().each { result += values[it] }
println result // Outputs 6You can learn more about closures in the Groovy documentation.
In Groovy, any gettersmethod that looks like get*() can also be accessed as a field (aka in property-style notation). For example, myFile.getName() is equivalent to myFile.name, myFile.getBaseName() is equivalent to myFile.baseName, and so on.
The transpose method is really useful when working with collections in Groovy.
It transposes the given lists. For example:
transpose([['a', 'b'], [1, 2]])- returns
[['a', 1], ['b', 2]]
- returns
transpose([['a', 'b', 'c']])- returns
[['a'], ['b'], ['c']]
- returns
The reason this is very useful in Nextflow is that many modules require inputs in the form of tuple val(meta), path(file)
In cases where you have to combine the output of many different tasks and amalgamate them into a single input for a process (E.g. Think about doing a lot a QC tasks on individual read files and then needing all those read files as input to perform a single genome assembly task), the input for that task might look like tuple val(metas), path(files). Notice how the basic input structure is still the same, just now there are a collection of meta maps and paths inside the collection.
So for example, let's say you have a process that outputs a channel that is something like this:
fastqFiles_ch
| view
/* Outputs
[[file: 1, meta: map], /path/to/file1.fastq]
[[file: 2, meta: map], /path/to/file2.fastq]
// The rest of the data (14 out of 16 [meta, path] collections not shown)
*/Then you can create a collection of collections with collect(flat: false):
fastqFiles_ch
| collect(flat: false)
| view
/* Outputs
[[[file: 1, meta: map], /path/to/file1.fastq], [[file: 2, meta: map], /path/to/file2.fastq], ... 14 out of 16 [meta, path] collections not shown]
*/And then you can split these collections into a metas collection and a files collection with Groovy's transpose method:
fastqFiles_ch
| collect(flat: false)
| map { it.transpose() }
| view
/* Outputs
[[[file: 1, meta: map], [file: 2, meta: map], ... 14 out of 16 metaMaps not shown], [/path/to/file1.fastq, /path/to/file2.fastq, ... 14 out of 16 file paths not shown]]
*/Now this channel is ready to get passed into a process that takes a tuple val(metas), path(files) input shape.
Note
The Groovy transpose method and the Nextflow transpose channel operator are two different things and will give you different results. Be aware of which one you need and when.
If you want to call a set method on every item in a Collection, Groovy provides this convenient "spread dot" notation
map { id, reads ->
reads.collect { it.getParent() }
}
// Is equivalent to
map { id, reads ->
reads*.getParent()
}You can significantly shorten lines if you combine property-style notation and spread-dot notation in groovy.
For example, the following 4 lines in the map are equivalent.
workflow {
Channel.fromFilePairs("data/reads/*/*_R{1,2}.fastq.gz")
| map { id, reads ->
reads.collect { it.getParent() }.collect { it.getName() }
reads.collect { it.parent }.collect { it.name }
reads*.getParent()*.getName()
reads*.parent*.name
}
| view
}There exists in Groovy and the JVM ecosystem a wealth of helper classes that can be imported into Nextflow scripts to help you accomplish some small tasks and helpful bits and pieces inside your Nextflow workflow.
For example, if you want to parse some JSON files in your code, you can import a Groovy JSON parser.
At the top of you main.nf file, add this:
import groovy.json.JsonSlurper
// We can also import a Yaml parser just as easily:
// import org.yaml.snakeyaml.Yaml
// new Yaml().load(new FileReader('your/data.yml'))You can then use the contents of this Groovy module in your Nextflow workflows:
def getFilteringResult(json_file) {
fastpResult = new JsonSlurper().parseText(json_file.text)
}
// ...
// In the workflow block
FASTP.out.json
| map { meta, json -> [meta, getFilteringResult(json)] }
| join( FASTP.out.reads )
| viewGroovy has a safe navigation operator ?.. It's used to safely access properties or methods of an object without throwing a NullPointerException if the object itself is null (Similar to optional chaining in JavaScript).
For example:
import groovy.json.JsonSlurper
def getFilteringResult(json_file) {
fastpResult = new JsonSlurper().parseText(json_file.text)
fastpResult?.summary?.after_filtering
}This code is safeguarded in case the summary or after_filtering objects don't exist. If these objects don't exist, the code will return early and return a null object.
A convienient way to perform a convient sanity-check on a Groovy expression is by using the Groovy web console or JDoodle.
An even better way to quickly check some Groovy or Nextflow code is by using the Nextflow console.
Activate the console by simply executing the following command in the terminal:
nextflow consoleThis will open a Nextflow REPL console for you to quickly test Groovy or Nextflow expressions.
- Press
command + Rto run the script in the console editor box. - Press
command + Wto clear the console output.
Here are some tips and shortcuts that I've found or came accross that don't really fit anywhere else in the notes.
There are a few preliminary steps you should do in your shell session before running a Nextflow pipeline on an HPC
-
Activate your conda environment containing Nextflow
-
Allocate a certain amount of memory for the Nextflow process. The Nextflow runtime runs on top of the Java virtual machine which, by design, tries to allocate as much memory as is available. This is not a good practice in HPC systems which are designed to share compute resources across many users and applications. To avoid this, specify the maximum amount of memory that can be used by the Java VM using the Java flags
-Xms(which sets the initial heap size) and-Xmx(which sets the maximum heap size). These can be specified using theNXF_OPTSenvironment variable.
Note
Different insitutions have different memory allowances. The KAUST IBEX cluster for example allows you to set -Xms3G and -Xmx5G, however for some infrastructures this may be too much. A lot of Nextflow and nf-core pipelines recommend using -Xms1G and -Xmx4G. Check what the allowances are to get the best performance on your runs.
- Change the
TMOUTenvironment variable. This variable controls the inactivity timeout for the shell (the default is 7200 seconds / 2 hours). When set, if a terminal session (including one inside tmux) remains idle for the specified duration, the shell will automatically terminate. It's useful to increase this because often times you'll run a process inside a tmux session, but when the process is complete you'll not be able to find your tmux session because it terminated 2 hours after the process completed, so you have to go back through the.nextflow.logfile to see if the process completed successfully instead of just viewing thestoutto the tmux session which is much cleaner.
conda activate "$(pwd)/env" && export NXF_OPTS='-Xms3G -Xmx5G' && echo "Java VM Heap memory allocated to a range of $(echo $NXF_OPTS | grep -oP '(?<=-Xms)\S+') and $(echo $NXF_OPTS | grep -oP '(?<=-Xmx)\S+') using the Nextflow ENV variable NXF_OPTS" && export TMOUT=172800 && echo "tmux timeout ENV variable (TMOUT) changed to $TMOUT seconds"Caution
Use updated command below. Need to finish notes about this.
conda activate "$(pwd)/env" && export NXF_OPTS='-Xms3G -Xmx5G' && echo "Java VM Heap memory allocated to a range of $(echo $NXF_OPTS | grep -oP '(?<=-Xms)\S+') and $(echo $NXF_OPTS | grep -oP '(?<=-Xmx)\S+') using the Nextflow ENV variable NXF_OPTS" && export TMOUT=172800 && echo "tmux timeout ENV variable (TMOUT) changed to $TMOUT seconds" && export SINGULARITY_CACHEDIR=$(pwd)/CACHE && export NXF_SINGULARITY_CACHEDIR=$(pwd)/NFCACHE && mkdir -p $SINGULARITY_CACHEDIR $NXF_SINGULARITY_CACHEDIR && echo "Created Singularity cache directory at $SINGULARITY_CACHEDIR" && echo "Created Nextflow Singularity cache directory at $NXF_SINGULARITY_CACHEDIR"tmux new-session -s SESSION_NAME -d 'source /PATH/TO/.bashrc ; source /PATH/TO/.bash_profile && conda activate CONDA_ENV export NXF_OPTS=NEXTFLOW_OPTIONS && echo "NXF_OPTS set to $NXF_OPTS" && export TMOUT=SECONDS_TO_TIMEOUT && echo "TMOUT set to $TMOUT" && nextflow run NEXTFLOW_PIPELINE -r PIPELINE_REVISION_NUMBER -c NEXTFLOW_CONFIGURATION -profile NEXTFLOW_PROFILES --outdir OUTPUT_DIRECTORY ; exec bash ' \; split-window -v 'sleep 20; tail -n 1000 -f .nextflow.log' \; attach (OPTIONAL)Example:
tmux new-session -s nfcore_rnaseq -d 'source /home/pampum/.bashrc ; source /home/pampum/.bash_profile && conda activate "$(pwd)/env" && export NXF_OPTS="-Xms3G -Xmx5G" && echo "NXF_OPTS set to $NXF_OPTS" && export TMOUT=172800 && echo "TMOUT set to $TMOUT" && nextflow run nf-core/rnaseq -r 3.14.0 -c nextflow.config -profile singularity,test --outdir results_rnaseq ; exec bash ' \; split-window -v 'sleep 20; tail -n 1000 -f .nextflow.log' \; attachExample without attaching:
tmux new-session -s nfcore_rnaseq -d 'source /home/pampum/.bashrc ; source /home/pampum/.bash_profile && conda activate "$(pwd)/env" && export NXF_OPTS="-Xms3G -Xmx5G" && echo "NXF_OPTS set to $NXF_OPTS" && export TMOUT=172800 && echo "TMOUT set to $TMOUT" && nextflow run nf-core/rnaseq -r 3.14.0 -c nextflow.config -profile singularity,test --outdir results_rnaseq ; exec bash ' \; split-window -v 'sleep 20; tail -n 1000 -f .nextflow.log'Breakdown:
-
tmux new-session -s nfcore_rnaseq -d:new-session: Creates a newtmuxsession.-s nfcore_rnaseq: Names the new sessionnfcore_rnaseq.-d: Starts the session in detached mode, meaning it will be created and run in the background, without immediately attaching you to it.
-
Session Command (
'...'):source /home/pampum/.bashrc: Sources the.bashrcfile, which is typically used for setting up the shell environment and environment variables.source /home/pampum/.bash_profile: Sources the.bash_profilefile, which is used for setting up the user environment, often executed for login shells.&& conda activate $(pwd)/env: Activates thecondaenvironment located at the current directory’senvsubdirectory.$(pwd)dynamically inserts the current directory path.&& export NXF_OPTS="-Xms3G -Xmx5G": Sets theNXF_OPTSenvironment variable to allocate a minimum of 3 GB (-Xms3G) and a maximum of 5 GB (-Xmx5G) of Java heap memory for the Nextflow process. This ensures that the process has adequate memory resources.&& echo "NXF_OPTS set to $NXF_OPTS": Outputs the value ofNXF_OPTSto confirm that it has been set correctly.&& export TMOUT=172800: Sets theTMOUTvariable to 172800 seconds (48 hours). This means that if the shell remains idle for 48 hours, it will automatically exit. This is useful for ensuring that idle sessions don’t remain open indefinitely.&& echo "TMOUT set to $TMOUT": Outputs the value ofTMOUTto confirm that it has been set.&& nextflow run nf-core/rnaseq -c nextflow.config -profile singularity,test --outdir results_rnaseq: Runs anextflowpipeline using thenf-core/rnaseqproject with the specified configuration file and profiles. The results will be saved toresults_rnaseq.; exec bash: Starts a new interactivebashshell after the command completes, keeping the terminal open for further interaction.
-
split-window -v:-v: Splits the current window vertically, creating a new pane below the existing one.
-
'sleep 20; tail -n 1000 -f .nextflow.log':sleep 20: Pauses for 20 seconds before executing the next command. This can give the primary command enough time to start and generate some output.tail -n 1000 -f .nextflow.log: Monitors the.nextflow.logfile in real-time, showing the last 1000 lines and updating as new lines are added. This is useful for tracking the log output of thenextflowrun.
-
attach:- Attaches to the
tmuxsession so that you can interact with it directly.
- Attaches to the
In addition, to monitor the jobs written by this command, you can use:
watch -d -n 1 squeue -u $USER -o \"%.8i %.90j %.8u %.2t %.10M %.6D %R\"Using the fetchngs nf-core pipeline
- Prepare your
ids.csvfile
PRJNA999857
PRJNA999856
PRJNA999855
...
- Collect the metadata. Use the
--skip_fastq_downloadflag
# Prepare your shell environment for a nextflow run
conda activate "$(pwd)/env" && export NXF_OPTS='-Xms3G -Xmx5G' && echo "Java VM Heap memory allocated to a range of $(echo $NXF_OPTS | grep -oP '(?<=-Xms)\S+') and $(echo $NXF_OPTS | grep -oP '(?<=-Xmx)\S+') using the Nextflow ENV variable NXF_OPTS" && export TMOUT=172800 && echo "tmux timeout ENV variable (TMOUT) changed to $TMOUT seconds"
# Run pipeline (only extracting metadata)
nextflow run nf-core/fetchngs -r 1.12.0 -c nextflow.config -profile mamba --input ids.csv --download_method sratools --outdir results --skip_fastq_download- Check the total size of all the
.fastqfiles that you would download
- If collecting paired end reads, the bytes of the paired end read files will be seperated with a
;on the resulting metadata table. Useawk -F '\t' '{split($29, a, ";"); sum += a[1] + a[2]} END {print sum}'to extract them.- Note: This command works regardless of if the reads are paired-end reads (i.e. regardless of if there is a
;in thefastq_bytescolumn). Can use it for single or paired end reads without breaking the program or affecting the accuracy of the calculation.
- Note: This command works regardless of if the reads are paired-end reads (i.e. regardless of if there is a
# Change into your results directory
cd results/metadata
# Check the total size of all your fastq files
ls -1 | xargs cat | sort | uniq | tail -n +2 | awk -F '\t' '{split($29, a, ";"); sum += a[1] + a[2]} END {print sum}' | numfmt --to=iec- If you have enough space, download all the data. Exclude the
--skip_fastq_downloadflag.
# Run pipeline (fetching fastq files)
nextflow run nf-core/fetchngs -r 1.12.0 -c nextflow.config -profile mamba --input ids.csv --download_method sratools --outdir results Can try to split up the ids.csv file into smaller chunks with the split command and then clear up the work directory between runs. The work directory will take up a lot of space and can cause issues.
# Split the ids.csv file into smaller chunks
split -n 3 --numeric-suffixes --additional-suffix=".csv" ids.csv ids.csv.
# Download the reads and clean up the working directory periodically
nextflow run nf-core/fetchngs -r 1.12.0 -c nextflow.config -profile mamba --input ids.csv.00.csv --download_method sratools --outdir results && \
rm -rf work && \
nextflow run nf-core/fetchngs -r 1.12.0 -c nextflow.config -profile mamba --input ids.csv.01.csv --download_method sratools --outdir results && \
rm -rf work && \
nextflow run nf-core/fetchngs -r 1.12.0 -c nextflow.config -profile mamba --input ids.csv.02.csv --download_method sratools --outdir results && \
rm -rf work