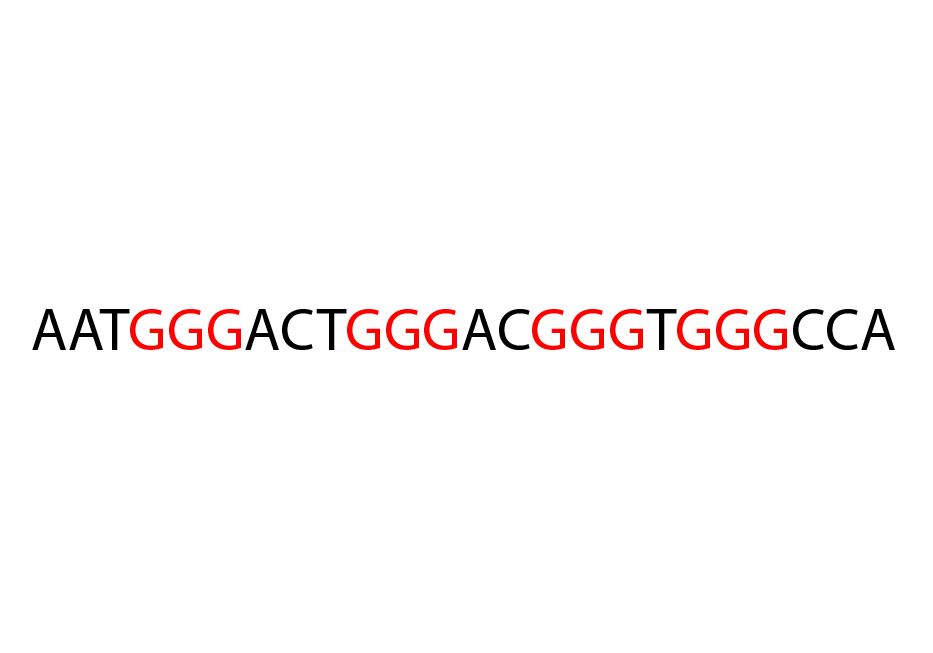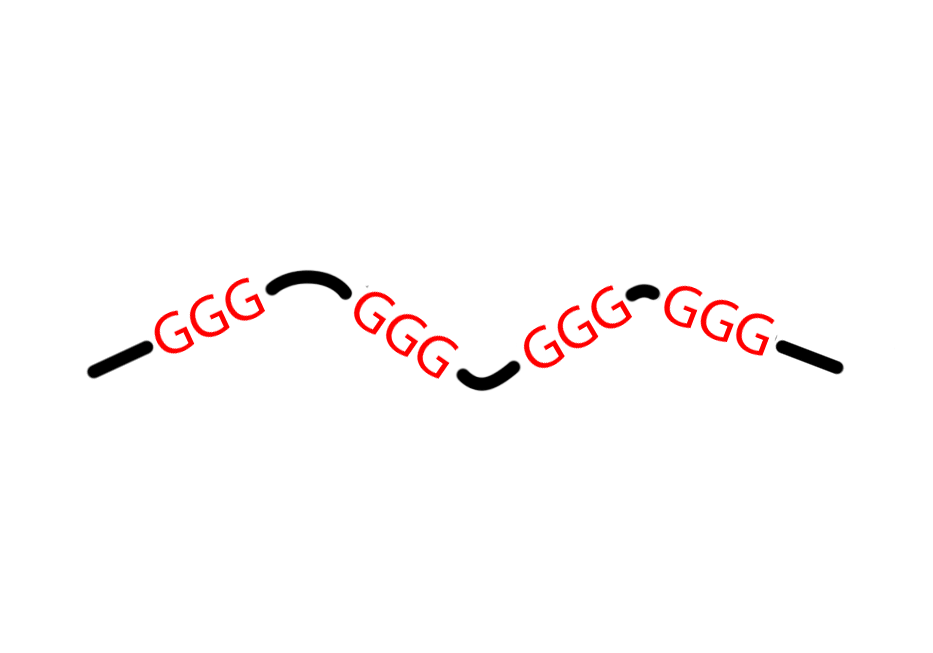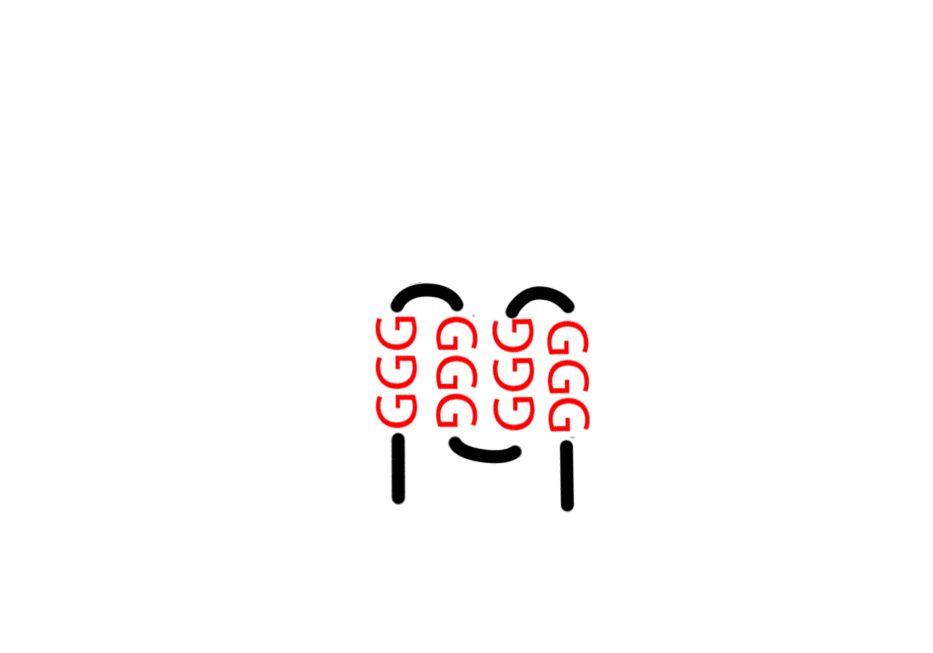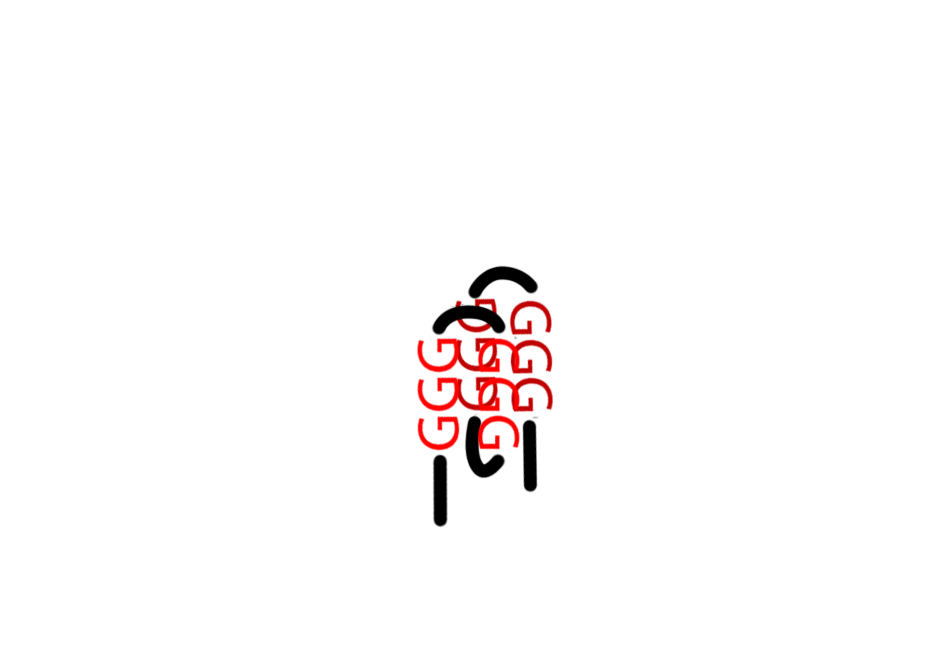Requires linux and the regex>=2016.3.2 module
TO INSTALL:
tar xf g4predict.tar.gz
cd g4predict
python setup.py build
python setup.py install
python setup.py test
usage: g4predict [-h] {intra,inter} ...
Predict putative G Quadruplexes using an extension of the Quadparser method;
NB: Output from g4predict is unlikely to be correctly sorted. use unix sort.
Author: Matthew Parker;
positional arguments:
{intra,inter}
intra Predict complete, intramolecular PG4s (i.e. PG4s which form
from one DNA/RNA strand). Uses the general pattern
G{x}([ATGC]{y,z}G{x}){3}.
inter Predict partial, intermolecular PG4s, which cannot form on
there own but might form with at least 1 other partial G4
from a different DNA/RNA molecule.
optional arguments:
-h, --help show this help message and exit
usage: g4predict intra [-h] -f FASTA -b BED [-t] [-s] [-F] [-M] [-c]
[-x TETRAD_SCORE_FACTOR] [-y LOOP_PEN_FACTOR]
[-tmin MIN_TETRAD] [-tmax MAX_TETRAD] [-lmin MIN_LOOP]
[-lmax MAX_LOOP] [-G ALLOW_G] [-B BULGES]
[-bmin MIN_BULGE] [-bmax MAX_BULGE]
[-z BULGE_PEN_FACTOR]
optional arguments:
-h, --help show this help message and exit
General:
-f FASTA, --fasta FASTA
Input fasta file, use '-' to read from stdin
-b BED, --bed BED Output bed file, use '-' to write to stdout
-t, --write-bed12 write bed12 output
-s, --write-bed6 write bed6 output instead of bed12 (some information
is lost)
-F, --filter-overlapping
use filtering method to remove overlapping PG4s,
yields the maximum number of high scoring, non-
overlapping PG4s
-M, --merge-overlapping
use merge method to flatten overlapping PG4s, output
is in bed6 and overrides the --write-bed12 flag
-c, --soft-mask if input fasta contains soft masking (i.e. lower case
nucleotides in repetitive or low complexity regions),
switch on case sensitivity to ignore these regions
Score:
Score method parameters. Scoring method is x*T - y*L - z*B, where T is the
number of tetrads, L is the total length of all loops and bulges, B is the
number of bulges, and x, y, and z are parameters which can be set by the
user.
-x TETRAD_SCORE_FACTOR, --tetrad-score-factor TETRAD_SCORE_FACTOR
factor to be multiplied by tetrad number in scoring
-y LOOP_PEN_FACTOR, --loop-pen-factor LOOP_PEN_FACTOR
factor to be multiplied by loop length in scoring
-z BULGE_PEN_FACTOR, --bulge-pen-factor BULGE_PEN_FACTOR
factor to be multiplied by bulge number in scoring
Tetrads:
-tmin MIN_TETRAD, --min-tetrad MIN_TETRAD
min number of tetrads allowed in predicted G4
-tmax MAX_TETRAD, --max-tetrad MAX_TETRAD
max number of tetrads in predicted G4
Loops:
-lmin MIN_LOOP, --min-loop MIN_LOOP
min loop length of predicted G4, either a single int
if all loops mins to the same length, or 3 comma
separated ints for loops 1 up to 3 (5'-> 3')
-lmax MAX_LOOP, --max-loop MAX_LOOP
max loop length of predicted G4, either a single int
if all loop maxes to be the same, or 3 comma separated
ints for loops 1 up to 3 (5'-> 3')
-G ALLOW_G, --allow-g ALLOW_G
allow G in PG4 loops, use 0 to disallow G in all
loops, or comma separated 0s or 1s to disallow G in
specific loops
Bulges (Intra Only):
-B BULGES, --bulges BULGES
how many bulges to allow in PG4s, can have up to one
per tetrad
-bmin MIN_BULGE, --min-bulge MIN_BULGE
min bulge length allowed in predicted G4
-bmax MAX_BULGE, --max-bulge MAX_BULGE
min bulge length allowed in predicted G4
usage: g4predict inter [-h] -f FASTA -b BED [-t] [-s] [-F] [-M] [-c]
[-x TETRAD_SCORE_FACTOR] [-y LOOP_PEN_FACTOR]
[-tmin MIN_TETRAD] [-tmax MAX_TETRAD] [-lmin MIN_LOOP]
[-lmax MAX_LOOP] [-G ALLOW_G] [-rmin MIN_G_RUNS]
[-rmax MAX_G_RUNS]
optional arguments:
-h, --help show this help message and exit
General:
-f FASTA, --fasta FASTA
Input fasta file, use '-' to read from stdin
-b BED, --bed BED Output bed file, use '-' to write to stdout
-t, --write-bed12 write bed12 output
-s, --write-bed6 write bed6 output instead of bed12 (some information
is lost)
-F, --filter-overlapping
use filtering method to remove overlapping PG4s,
yields the maximum number of high scoring, non-
overlapping PG4s
-M, --merge-overlapping
use merge method to flatten overlapping PG4s, output
is in bed6 and overrides the --write-bed12 flag
-c, --soft-mask if input fasta contains soft masking (i.e. lower case
nucleotides in repetitive or low complexity regions),
switch on case sensitivity to ignore these regions
Score:
Score method parameters. Scoring method is x*T - y*L - z*B, where T is the
number of tetrads, L is the total length of all loops and bulges, B is the
number of bulges, and x, y, and z are parameters which can be set by the
user.
-x TETRAD_SCORE_FACTOR, --tetrad-score-factor TETRAD_SCORE_FACTOR
factor to be multiplied by tetrad number in scoring
-y LOOP_PEN_FACTOR, --loop-pen-factor LOOP_PEN_FACTOR
factor to be multiplied by loop length in scoring
Tetrads:
-tmin MIN_TETRAD, --min-tetrad MIN_TETRAD
min number of tetrads allowed in predicted G4
-tmax MAX_TETRAD, --max-tetrad MAX_TETRAD
max number of tetrads in predicted G4
Loops:
-lmin MIN_LOOP, --min-loop MIN_LOOP
min loop length of predicted G4, either a single int
if all loops mins to the same length, or 3 comma
separated ints for loops 1 up to 3 (5'-> 3')
-lmax MAX_LOOP, --max-loop MAX_LOOP
max loop length of predicted G4, either a single int
if all loop maxes to be the same, or 3 comma separated
ints for loops 1 up to 3 (5'-> 3')
-G ALLOW_G, --allow-g ALLOW_G
allow G in PG4 loops, use 0 to disallow G in all
loops, or comma separated 0s or 1s to disallow G in
specific loops
Inter:
-rmin MIN_G_RUNS, --min-g-runs MIN_G_RUNS
min runs of G to use to predict partial PG4s
-rmax MAX_G_RUNS, --max-g-runs MAX_G_RUNS
max runs of G to use to predict partial PG4s