To use this it is probably best to load the anaconda enviorment on the cluster
#!/usr/bin/env bash
unset PYTHONPATH
module purge
. /etc/profile.d/modules.sh
module load modules modules-init modules-gs/prod modules-eichler
module load anaconda/201710
or just
source conda.cfg
This seems to need miniconda/4.5.12
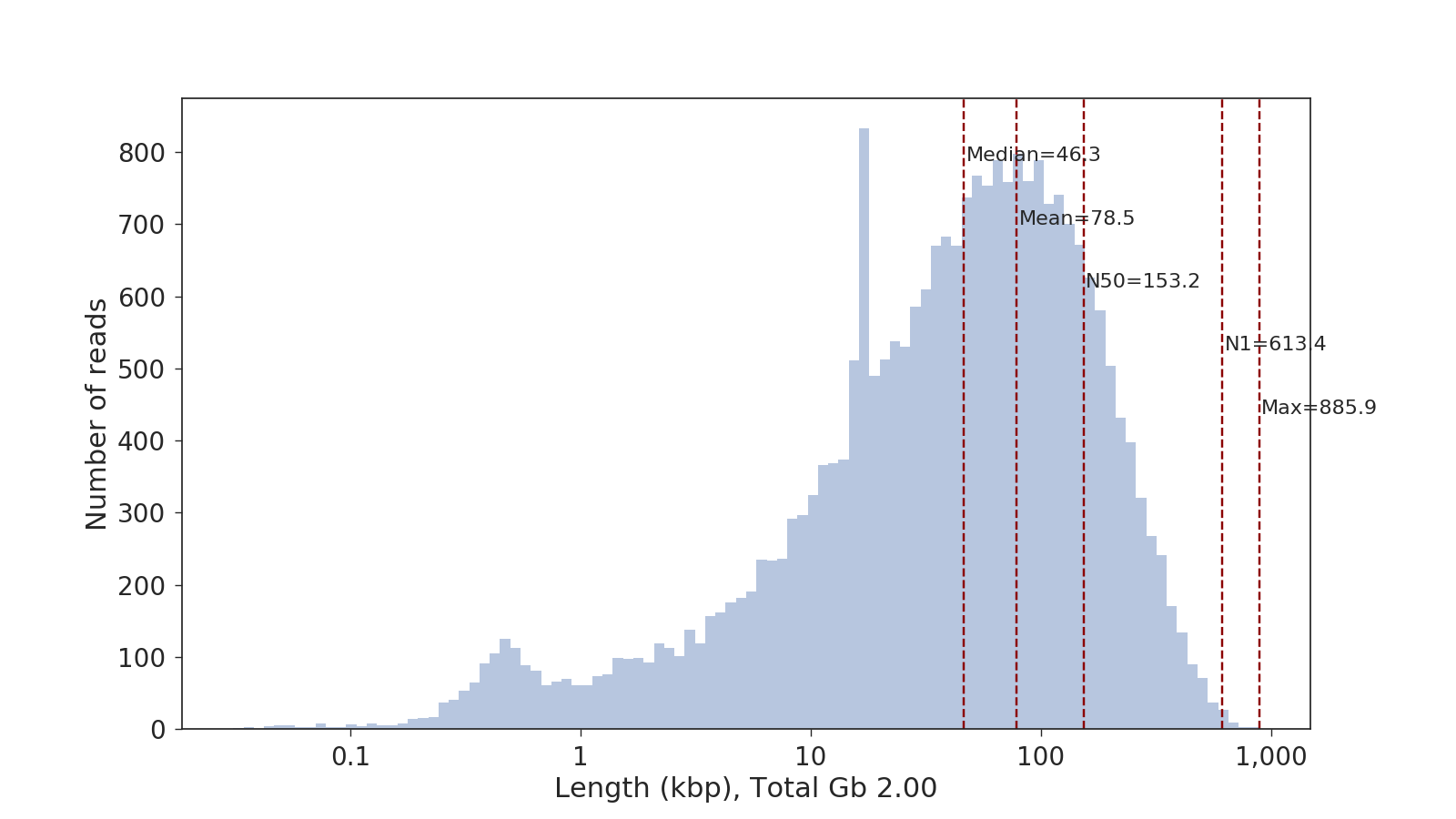
SeqLengths.py --help
usage: SeqLengths.py [-h] [-f FOFN] [-o OUT] [-p PLOT] [--plotwith PLOTWITH]
[-t THREADS]
[infiles [infiles ...]]
Get the lengths of reads from fasta/fastq/(.gz) files.
positional arguments:
infiles input fasta/fastq file
optional arguments:
-h, --help show this help message and exit
-f FOFN, --fofn FOFN input FOFN of fasta/fastq files, replaces infiles
argument
-o OUT, --out OUT output lengths file. Has the form: readname, length
-p PLOT, --plot PLOT Specify a place to plot a hsitogram of read lengths
--plotwith PLOTWITH If you have already calcualted readlength before you
can specify plotwith to avoid rereading the fasta
files
-t THREADS, --threads THREADS
number of threads to use, only used in reading in
fasta/fastq files