This is the official code for "Robust Prototypical Few-Shot Organ Segmentation with Regularized Neural-ODEs", IEEE Transactions of Medical Imaging, 2022.
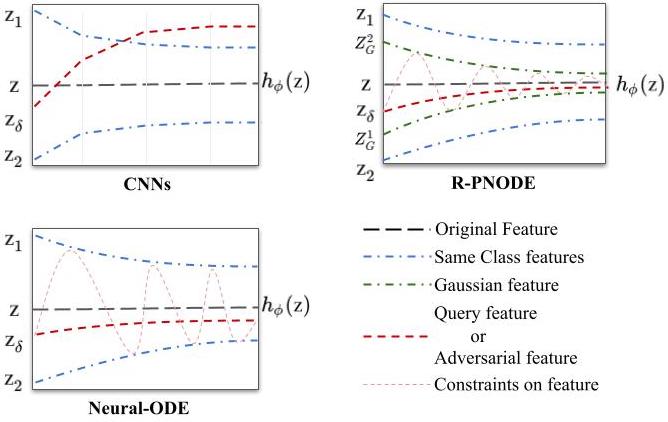
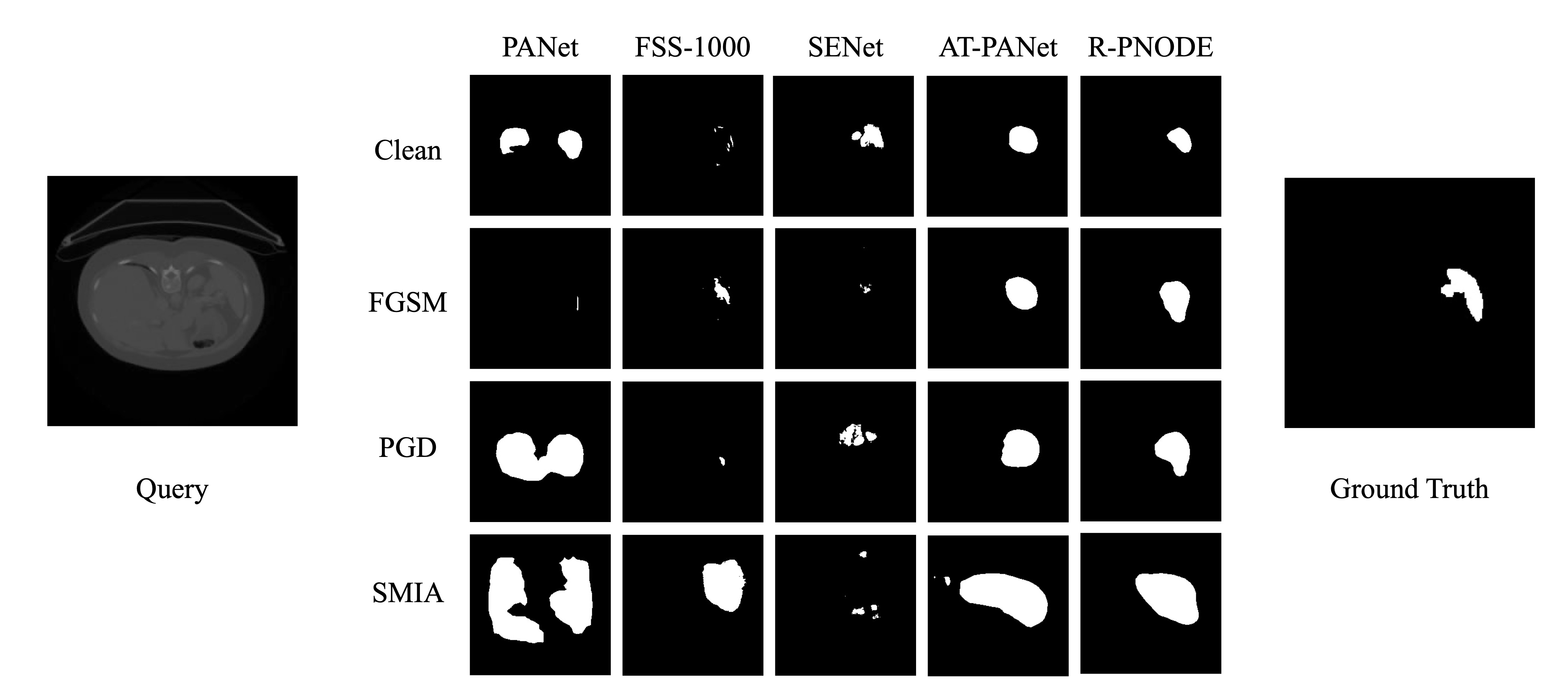
Despite the tremendous progress made by deep learning models in image semantic segmentation, they typically require large annotated examples, and increasing attention is being diverted to problem settings like Few-Shot Learning (FSL) where only a small amount of annotation is needed for generalisation to novel classes. This is especially seen in medical domains where dense pixel-level annotations are expensive to obtain. In this paper, we propose Regularized Prototypical Neural Ordinary Differential Equation (R-PNODE), a method that leverages intrinsic properties of Neural-ODEs, assisted and enhanced by additional cluster and consistency losses to perform Few-Shot Segmentation (FSS) of organs. R-PNODE constrains support and query features from the same classes to lie closer in the representation space thereby improving the performance over the existing Convolutional Neural Network (CNN) based FSS methods. We further demonstrate that while many existing Deep CNN-based methods tend to be extremely vulnerable to adversarial attacks, R-PNODE exhibits increased adversarial robustness for a wide array of these attacks. We experiment with three publicly available multi-organ segmentation datasets in both in-domain and cross-domain FSS settings to demonstrate the efficacy of our method. In addition, we perform experiments with seven commonly used adversarial attacks in various settings to demonstrate R-PNODE's robustness. R-PNODE outperforms the baselines for FSS by significant margins and also shows superior performance for a wide array of attacks varying in intensity and design.
To install this repository and its dependent packages, run the following.
git clone https://github.com/rpnode-fss/RPNODE_FSS.git
cd R-PNODE
conda create --name R-PNODE # (optional, for making a conda environment)
pip install -r requirements.txt
The processed datasets can be downloaded from here.
Some relevant trained model weights can be downloaded from here.
Change the paths to BCV, CT-ORG and Decathlon datasets in config.py and test_config.py according to paths on your local. Also change the path to ImageNet pretrained VGG model weights in these files.
To train R-PNODE, run
python3 train.py with dataset=BCV model_name=<save-name> target=<train-target> n_shot=<shot> ode_layers=3 ode_time=4
Further parameters like the standard deviation of gaussian perturbation can be changed in the training config.
So, for example, to train the experiment BCV 1-shot with Spleen organ as the novel class, the command would be
python3 train.py with dataset=BCV model_name=bcv_1shot_spleen target=1 n_shot=1 ode_layers=3 ode_time=4
This will store model weights with the name bcv_1shot_spleen_tar1.pth in the model root directory. Please refer to the class mapping below to find which target index to use for which target class. Note that a single model is needed to test the method in both in-domain and cross-domain settings for a particular shot and target. Similarly, all different attacks are tested on a single trained model.
To test a trained model, run
python3 test_attacked.py with snapshot=<weights-path> target=<test-target> dataset=<BCV/CTORG/Decathlon> attack=<Clean/FGSM/PGD/SMIA> attack_eps=<eps> to_attack=<q/s>
Arguments for some particular settings are:
| Setting | Arguments |
|---|---|
| BCV in-domain 1-shot Liver | dataset=BCV n_shot=1 target=6 |
| BCV in-domain 3-shot Spleen | dataset=BCV n_shot=3 target=1 |
| BCV -> CT-ORG cross-domain 1-shot Liver | dataset=CTORG n_shot=1 target=1 |
| BCV -> Decathlon cross-domain 3-shot Liver | dataset=Decathlon n_shot=3 target=2 |
| BCV -> Decathlon cross-domain 1-shot Spleen | dataset=Decathlon n_shot=1 target=6 |
Note particularly for the cross-domain settings that the target class index used during training may be different from that used during testing. Special care must be taken that models trained for particular target organs are tested for the same organs to avoid misleading results. Please refer to the class mapping at the end of the readme for exact target indices.
The possible options for the attack argument are:
- clean (standard FSS, without any attack)
- fgsm
- pgd
- smia
- bim
- cw
- dag
- auto
These are case insensitive, and using variants like FGSM, FGsm, fGsM will also lead to same effect.
This command can be used for testing on all settings, namely 1-shot and 3-shot, liver and spleen and Clean, FGSM, PGD, SMIA, BIM, CW, DAG and Auto-Attack with different epsilons.
Visualization can be enabled by setting save_vis as True. The path where the visualisations will be saved can be modified here.
This repository adapts from BiGRU-FSS. 4 of the baselines are from here. The other baseline is the Neural-ODE based SONet. Attacks are adapted from well known libraries like CleverHans and TorchAttacks. Other attacks are adapted from their respective sources: SMIA, DAG and Auto-attack. We thank the authors for their awesome works.
BCV:
Liver: 6
Spleen: 1
CT-ORG:
Liver: 1
Decathlon:
Liver: 2
Spleen: 6