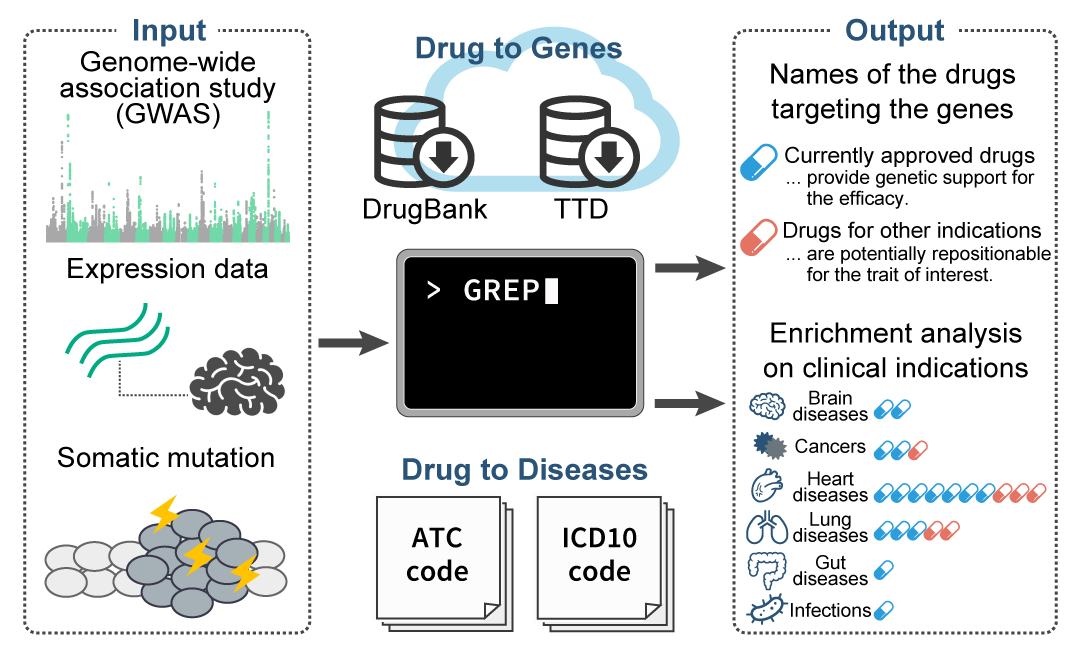
GREP can quantify an enrichment of the user-defined set of genes in the target of clinical indication categories and capture potentially repositionable drugs targeting the gene set. Both can be run in a few seconds!
The user input is a gene list from any source of genomic studies. GREP tells you (i) what kind of disease categories are pharmaco-genetically associated with the gene set and (ii) what kind of medications can have a potential for being repositioned to another indication.
Our paper is out!!!
Sakaue S and Okada Y. GREP: Genome for REPositioning drugs. Bioinformatics. 2019
Please cite this paper if you use GREP or any material in this repository.
GREP is a command line python software, and the following libraries are required.
- scipy
- argparse
- numpy
- pandas
This software is compatible with both versions of python: 2 and 3.
In order to get started with GREP, you can clone this repo by the following commands,
$ git clone https://github.com/saorisakaue/GREP
$ cd ./GREP
Or, you can also use pip to install
currently under development
An example basic command is as follows;
$ python grep.py --genelist ./example/megastroke.genes --out my_GREP_test --test ATC --output-drug-nameMake a text file with one column, which contains genes by HGNC gene symbol.
Please have a look at ./example/ directory for example gene sets.
./example/megastroke.genescan be used as genes identified by MEGASTROKE consortium (Nat Genet 2018)../example/RA_trans.genescan be used as genes identified in RA meta-analysis by Okada et al (Nature 2014).
If your would like to perform post-GWAS analysis, and if you would like to convert SNP-level statisitcs to gene-level ones, here are some of the options.
- A method used in MAGENTA. Reimplementation in python for the conversion is distributed here. Acknowledgement goes to Masa Kanai.
- MAGMA (external link)
- PASCAL (external link)
To make a gene set, define P value threshold of your choice, and make a text file with a set of genes which pass that threshold using the output from the above softwares.
| Option name | Descriptions | Required | Default |
|---|---|---|---|
--genelist, -g |
Input your list of genes as a text file with one column. | Yes | None |
--out, -o |
An output prefix. | Yes | None |
--test, -t |
Choose from ATC or ICD for a drug indication categorization system. |
Yes | None |
--output-drug-name, -d |
If you want to know the drug names targeting your genes, set this flag (without any arguments after that). | No | False |
--background, -b |
A list of all genes in the scope of your analysis if available. This will be used as background genes. | No | All the genes targeted by the drugs in the database |
- ATC; The Anatomical Therapeutic Chemical (ATC) Classification System is used for the classification of active ingredients of drugs according to the organ or system on which they act and their therapeutic, pharmacological and chemical properties. Large annotation has 14 anatomical categories, which are further categorized into ~90 detailed classes in total. Drug-category information was retrieved from KEGG DRUG website.
- ICD; The 10th revision of the International Statistical Classification of Diseases and Related Health Problems categorizes diseases into 26 large groups, which are further subdivided into ~ 200 classes. Drug-indicated disease category information was retrieved from TTD website.
The above command generates two text files for ATC analysis (tests for large categories and detailed categories), and one file for ICD analysis with the first line being a header.
Below is an example output from ATC analysis for large categories.
$ head my_GREP_test.ATC.large.txt
#Group GroupName OddsRatio FisherExactP TargetGene:DrugNames
A ALIMENTARY TRACT AND METABOLISM 0.8160667251975418 0.7076001488153763 ACACB:metformin;TRPV4:citric acid
B BLOOD AND BLOOD FORMING ORGANS 5.287042099507928 0.004721436181224572 F11:conestat alfa;FGA:alteplase,anistreplase,reteplase,tenecteplase;KLKB1:ecallantide,conestat alfa;PDE3A:cilostazol;PLG:streptokinase,alteplase,anistreplase,urokinase,reteplase,tenecteplase,melagatran,aminocaproic acid,tranexamic acid
C CARDIOVASCULAR SYSTEM 1.4418238993710693 0.327034895762871 ATP2A2:gallopamil;EDNRA:bosentan,ambrisentan,sitaxentan,macitentan;PDE3A:amrinone,milrinone,enoximone,trapidil;PLG:ranolazine| Column | Column Name | Descriptions |
|---|---|---|
| 1 | Group | An alphabetical code for each category. |
| 2 | GroupName | A description of the anatomical category. |
| 3 | OddsRatio | The enrichment odds ratio quantifying the degree of the overlap between genes included in the input and genes targeted by the drugs included in the category tested. |
| 4 | FisherExactP | The enrichment P value by Fisher's exact test (one-sided). |
| 5 | TargetGene:DrugNames | The overpapped genes and the drugs targeting those genes (if any). |
This software is freely available for academic users. Usage for commercial purposes is not allowed. Please refer to the LICENCE page.
We thank Masa Kanai for helping us refactor the software implementation.
Any questions? Saori Sakaue (ssakaue[at]sg.med.osaka-u.ac.jp)