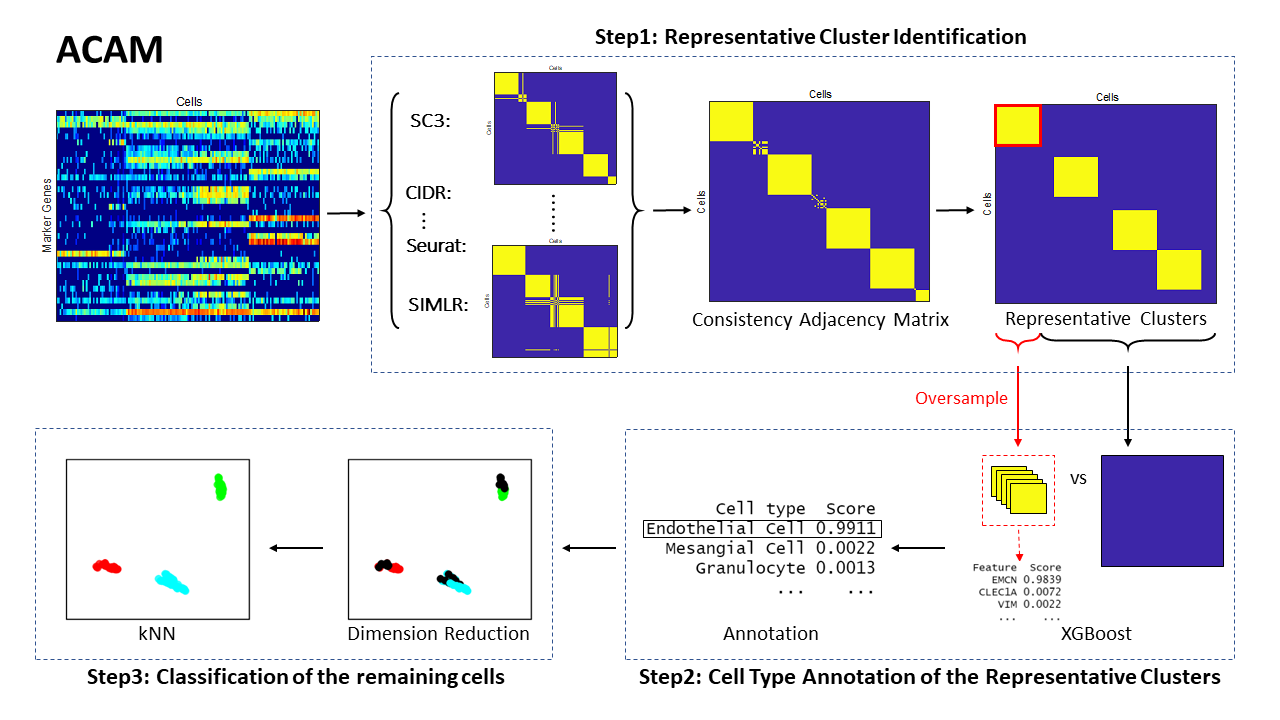
Automatic Cell type Annotation Method (ACAM) is proposed based on marker genes' information. This method first finds the representative clusters by searching for the consistent subgroups across the results of several popular clustering methods. Such technique guarantees that the cells in the same cluster have very high probabilities of being from the same cell type. Then by selecting the features that discriminate one cluster from all the remaining cells, the potential marker genes are identified. The cell types are determined by defining a cell type importance score to match these marker genes with the validated ones. For those cells that do not belong to any of these clusters, we use
You can use our method ACAM by installing this R package with the following code.
devtools::install_github("yuc0824/ACAM")
ACAM requires the following inputs.
DFThe cell-by-gene expression matrix with rows of cells and column of marker genes.gene.markersThe species- and tissue-specific marker genes obtained from CellMatch database. You can define your own markers.
We put Chen dataset in this package as the example.
DF_chen: The dataset Chen obtained from GEO with the accession number: GSE99701.
GM_chen: The species- and tissue-specific marker genes obtained from the database CellMatch.
Before putting into ACAM, make sure that the dataset are log-normalized.
library(ACAM)
data("DF_chen")
data("GM_chen")
First, we get the clustering results from four of the five popular clustering methods with max pairwise ARI variances.
cluster_results <- ACAM_cluster(DF = DF_chen)
The dataset is then annotated by the function below.
annotation_results <- ACAM_annotation(DF = DF_chen, cluster_results = cluster_results, gene.markers = GM_chen)
All the data, results and visualizations in the paper can be found at the repository https://github.com/yuc0824/ACAMdata.
Yu Chen, Fudan University
Shuqin Zhang, Fudan University