Fastq2fasta converts single or paired end fastQ in a single fasta file for Blastn input.
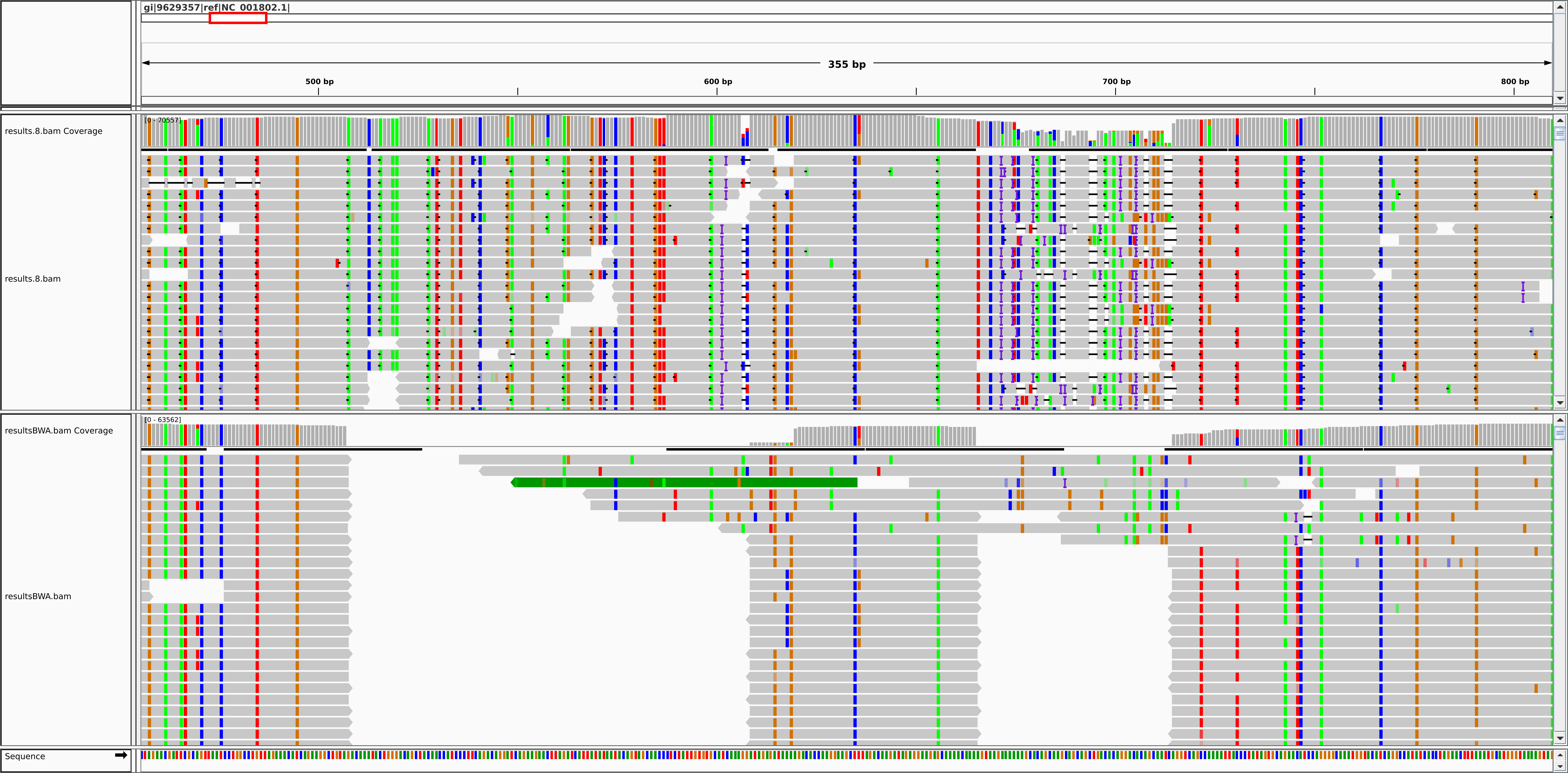
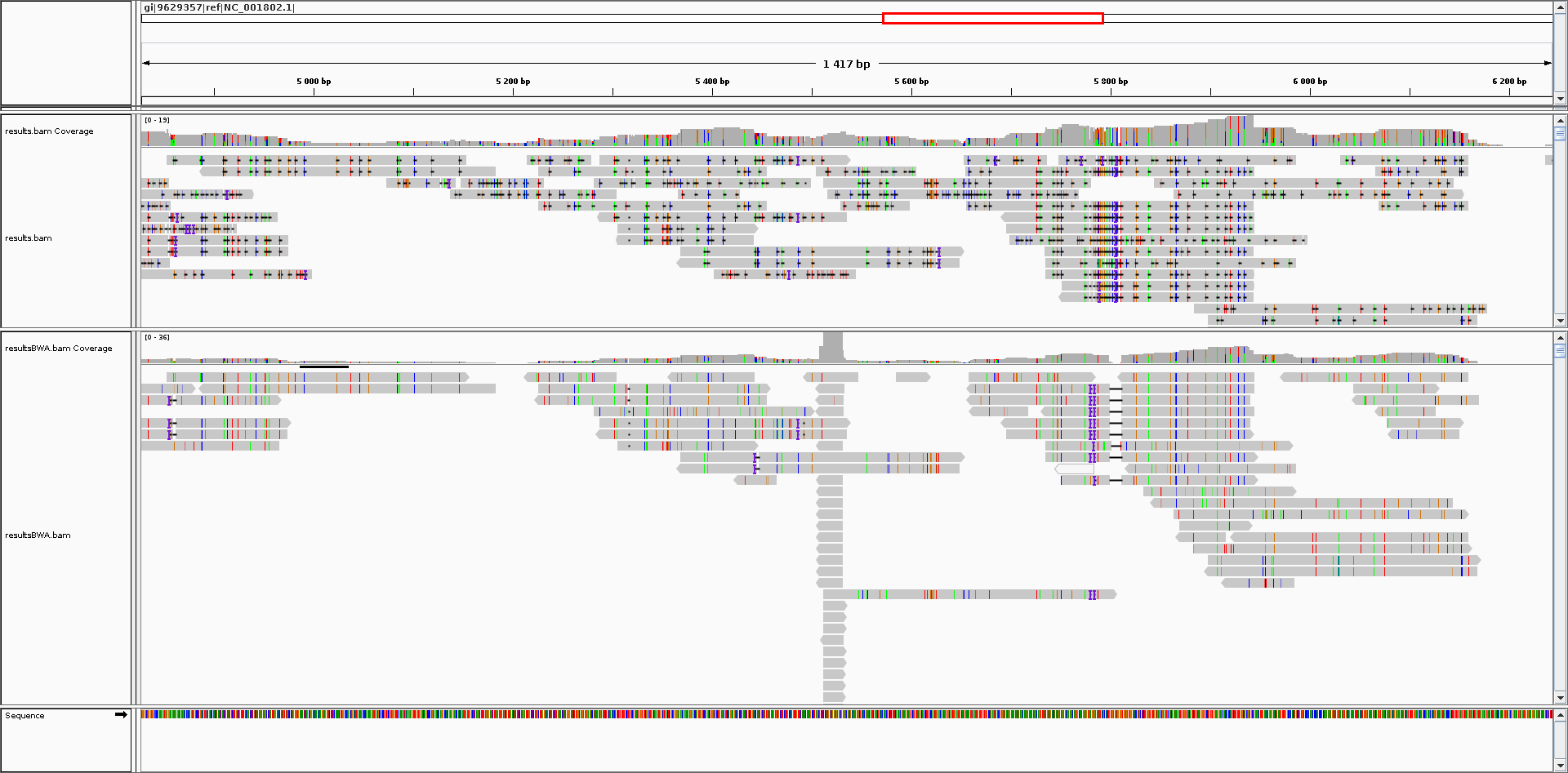
Blast2Bam uses the XML results of Blastn, the reference and the fastQ or fasta file(s) to output a SAM file.
In case of paired end data, Blast results from the first and second fastQ/fasta are paired in the SAM output.
The file generated by Blast2Bam is compatible with SAMtools and pass the picard-tools ValidateSamFile test (when the secondary alignments are removed).
- gcc
- libxml2
- xsltproc
- zlib
$ make
$ fastq2fasta [options] FastQ_1 [FastQ_2] > out.fasta$ blast2bam [options] blast.xml ref.fasta FastQ_1 [FastQ_2] > out.sam| Option | Description |
|---|---|
| -o FILE | File output. Default: stdout. |
| -h | Help |
| Option | Description |
|---|---|
| -o FILE | File output. Default: stdout. |
| -p | Interleaved. The input fastQ/fasta is an interleaved list of sequences forward and reverse. |
| -R STR | Read group header line. Should look like this : `@RG\tID:foo\t[...]`. |
| -W INT | Minimum alignment length. The alignment is displayed only if its length is greater than INT. Default: minimum length calculated based on read length. |
| -c | Short version of the CIGAR string ('M' instead of '=' and 'X'). |
| -z | Adjusted position. The position of the alignment is adjusted to the position of the reference. |
| -h | Help |
.SHELL = /bin/bash
BINDIR = ../../bin/
SRCDIR = ../../src/
FASTQ = test_1.fastq.gz test_2.fastq.gz
DB = hiv.fasta
results.sam: $(addprefix ${BINDIR}, blast2bam fastq2fasta) ${FASTQ} $(addsuffix .nin, ${DB})
$(addprefix ${BINDIR}, fastq2fasta) ${FASTQ} | \
blastn -db ${DB} -num_threads 4 -word_size 8 -outfmt 5 | \
$(addprefix ${BINDIR}, blast2bam) -o $@ -R '@RG\tID:foo\tSM:sample' - ${DB} ${FASTQ}
$(addprefix ${BINDIR}, blast2bam fastq2fasta):
(cd ${SRCDIR}; ${MAKE})
$(addsuffix .nin,${DB}): ${DB}
makeblastdb -in $< -dbtype 'nucl'(...)
@ERR656485.2 2 length=300
NTGGGCTAAAGGCCTTTTCCTCTATTACTTTTACCCATGCATTTAAAGTTCTAGGTGACATGGCCTGGTGTACCATTTGCCCTTGGAGATTTTGCACTATAGGATAATTTTGACTGACCTTCCCGTCAGCCCCTTTTTCCTGCTGTGTCTTTTGCTGACTTTGCTGACATTTGTTTTGTTCTTCCTCTATCTTAGATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGATAGAGGATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAAGAAGAAAAGCAAGCAACACTAGG
+ERR656485.2 2 length=300
!8ACC@FGGGGGGGGFGGGGGGGGGGGGGGGGGGCFGGGGGGGGGGGFGGGGFCEFFGGGGGGGGGGGFGECFFGDGGGGGGGGGGEFGGGEGGGEFGGGGGGGGEFFFGGE?FFGGGGGFFGGGGGGFGGEGGGGGFGGGGGGGGGGFGGCEGDGGGGGGGGFGGGG9EGFFEGGGGGGGGGGGFFGF;DEGGGGFGGG>868EFGGFD?FGFGG55:FDFFF>:95??A=FAFF==B=46B47).1592::EEB?25-9@CE5>=/6>E02>96>(-.((.((,(((((,((((.,)(
(...)
(...)
@ERR656485.2 2 length=300
NAGATAGAGGAAGAACAAAACAAATGTCAGCAAAGTCAGCAAAAGACACAGCAGGAAAAAGGGGCTGACGGGAAGGTCAGTCAAAATTATCCTATAGTGCAAAATCTCCAAGGGCAAATGGTACACCAGGCCATGTCACCTAGAACTTTAAATGCATGGGTAAAAGTAATAGAGGAAAAGGCCTTTAGCCCAGAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTCTCATTACAAAAAAAACATACACAATAAATGATATAAGCGGAATCAACAGCATGA
+ERR656485.2 2 length=300
!8A@CGGEFGFGCDFGGGGGGGGGGGGGGFGGGGGFGFGGGGGGGGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGFGGFGGGGGGGGGEGFGFGGGFFGGGGGGGGFGGGGGGGGGGGGFFFFGGGGGG=FFGGFFDGGGGGGGG8FGFGGGGGGGGGFGGGGGGGGGGFDGGFGGFGGGFFFGFF8DFDFDFFFFFFFFFBCDB<@EAFB@ABAC@CDFF?4>EEFE<*>BDAFB@FFBFF>((6<5CC.;C;=D9106(.))).)-46<<))))))))))((,(-)))()((()))
(...)
(...)
<Iteration>
<Iteration_iter-num>3</Iteration_iter-num>
<Iteration_query-ID>Query_3</Iteration_query-ID>
<Iteration_query-def>ERR656485.2</Iteration_query-def>
<Iteration_query-len>300</Iteration_query-len>
<Iteration_hits>
<Hit>
<Hit_num>1</Hit_num>
<Hit_id>gnl|BL_ORD_ID|0</Hit_id>
<Hit_def>gi|9629357|ref|NC_001802.1| Human immunodeficiency virus 1, complete genome</Hit_def>
<Hit_accession>0</Hit_accession>
<Hit_len>9181</Hit_len>
<Hit_hsps>
<Hsp>
<Hsp_num>1</Hsp_num>
<Hsp_bit-score>148.852</Hsp_bit-score>
<Hsp_score>80</Hsp_score>
<Hsp_evalue>4.07002e-39</Hsp_evalue>
<Hsp_query-from>2</Hsp_query-from>
<Hsp_query-to>120</Hsp_query-to>
<Hsp_hit-from>833</Hsp_hit-from>
<Hsp_hit-to>715</Hsp_hit-to>
<Hsp_query-frame>1</Hsp_query-frame>
<Hsp_hit-frame>-1</Hsp_hit-frame>
<Hsp_identity>106</Hsp_identity>
<Hsp_positive>106</Hsp_positive>
<Hsp_gaps>0</Hsp_gaps>
<Hsp_align-len>119</Hsp_align-len>
<Hsp_qseq>TGGGCTAAAGGCCTTTTCCTCTATTACTTTTACCCATGCATTTAAAGTTCTAGGTGACATGGCCTGGTGTACCATTTGCCCTTGGAGATTTTGCACTATAGGATAATTTTGACTGACCT</Hsp_qseq>
<Hsp_hseq>TGGGCTGAAAGCCTTCTCTTCTACTACTTTTACCCATGCATTTAAAGTTCTAGGTGATATGGCCTGATGTACCATTTGCCCCTGGATGTTCTGCACTATAGGGTAATTTTGGCTGACCT</Hsp_hseq>
<Hsp_midline>|||||| || ||||| || |||| ||||||||||||||||||||||||||||||||| |||||||| |||||||||||||| |||| || ||||||||||| |||||||| |||||||</Hsp_midline>
</Hsp>
(...)(...)
<Iteration>
<Iteration_iter-num>4</Iteration_iter-num>
<Iteration_query-ID>Query_4</Iteration_query-ID>
<Iteration_query-def>ERR656485.2</Iteration_query-def>
<Iteration_query-len>300</Iteration_query-len>
<Iteration_hits>
<Hit>
<Hit_num>1</Hit_num>
<Hit_id>gnl|BL_ORD_ID|0</Hit_id>
<Hit_def>gi|9629357|ref|NC_001802.1| Human immunodeficiency virus 1, complete genome</Hit_def>
<Hit_accession>0</Hit_accession>
<Hit_len>9181</Hit_len>
<Hit_hsps>
<Hsp>
<Hsp_num>1</Hsp_num>
<Hsp_bit-score>152.546</Hsp_bit-score>
<Hsp_score>82</Hsp_score>
<Hsp_evalue>3.14632e-40</Hsp_evalue>
<Hsp_query-from>74</Hsp_query-from>
<Hsp_query-to>194</Hsp_query-to>
<Hsp_hit-from>715</Hsp_hit-from>
<Hsp_hit-to>835</Hsp_hit-to>
<Hsp_query-frame>1</Hsp_query-frame>
<Hsp_hit-frame>1</Hsp_hit-frame>
<Hsp_identity>108</Hsp_identity>
<Hsp_positive>108</Hsp_positive>
<Hsp_gaps>0</Hsp_gaps>
<Hsp_align-len>121</Hsp_align-len>
<Hsp_qseq>AGGTCAGTCAAAATTATCCTATAGTGCAAAATCTCCAAGGGCAAATGGTACACCAGGCCATGTCACCTAGAACTTTAAATGCATGGGTAAAAGTAATAGAGGAAAAGGCCTTTAGCCCAGA</Hsp_qseq>
<Hsp_hseq>AGGTCAGCCAAAATTACCCTATAGTGCAGAACATCCAGGGGCAAATGGTACATCAGGCCATATCACCTAGAACTTTAAATGCATGGGTAAAAGTAGTAGAAGAGAAGGCTTTCAGCCCAGA</Hsp_hseq>
<Hsp_midline>||||||| |||||||| ||||||||||| || |||| |||||||||||||| |||||||| ||||||||||||||||||||||||||||||||| |||| || ||||| || ||||||||</Hsp_midline>
</Hsp>
(...)@SQ SN:gi|9629357|ref|NC_001802.1| LN:9181
@RG ID:foo SM:sample
@PG ID:Blast2Bam PN:Blast2Bam VN:0.1 CL:../../bin/blast2bam -o results.sam -R @RG\tID:foo\tSM:sample - hiv.fasta test_1.fastq.gz test_2.fastq.gz
(...)
ERR656485.2 83 gi|9629357|ref|NC_001802.1| 715 60 180S7=1X8=1X11=1X2=2X4=1X14=1X8=1X33=1X4=1X2=1X5=1X2=1X6=1S = 715 -119 CCTAGTGTTGCTTGCTTTTCTTCTTTTTTTTTTCAAGCAGAAGACGGCATACGAGATCCTCTATCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCTAAGATAGAGGAAGAACAAAACAAATGTCAGCAAAGTCAGCAAAAGACACAGCAGGAAAAAGGGGCTGACGGGAAGGTCAGTCAAAATTATCCTATAGTGCAAAATCTCCAAGGGCAAATGGTACACCAGGCCATGTCACCTAGAACTTTAAATGCATGGGTAAAAGTAATAGAGGAAAAGGCCTTTAGCCCAN (),.((((,(((((,((.((.-(>69>20E>6/=>5EC@9-52?BEE::2951.)74B64=B==FFAF=A??59:>FFFDF:55GGFGF?DFGGFE868>GGGFGGGGED;FGFFGGGGGGGGGGGEFFGE9GGGGFGGGGGGGGDGECGGFGGGGGGGGGGFGGGGGEGGFGGGGGGFFGGGGGFF?EGGFFFEGGGGGGGGFEGGGEGGGFEGGGGGGGGGGDGFFCEGFGGGGGGGGGGGFFECFGGGGFGGGGGGGGGGGFCGGGGGGGGGGGGGGGGGGFGGGGGGGGF@CCA8! NM:i:13 RG:Z:foo AS:i:80 XB:f:148.852 XE:Z:4.07e-39
ERR656485.2 163 gi|9629357|ref|NC_001802.1| 715 60 73S7=1X8=1X11=1X2=2X4=1X14=1X8=1X33=1X4=1X2=1X5=1X2=1X8=106S = 715 119 NAGATAGAGGAAGAACAAAACAAATGTCAGCAAAGTCAGCAAAAGACACAGCAGGAAAAAGGGGCTGACGGGAAGGTCAGTCAAAATTATCCTATAGTGCAAAATCTCCAAGGGCAAATGGTACACCAGGCCATGTCACCTAGAACTTTAAATGCATGGGTAAAAGTAATAGAGGAAAAGGCCTTTAGCCCAGAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTCTCATTACAAAAAAAACATACACAATAAATGATATAAGCGGAATCAACAGCATGA !8A@CGGEFGFGCDFGGGGGGGGGGGGGGFGGGGGFGFGGGGGGGGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGFGGFGGGGGGGGGEGFGFGGGFFGGGGGGGGFGGGGGGGGGGGGFFFFGGGGGG=FFGGFFDGGGGGGGG8FGFGGGGGGGGGFGGGGGGGGGGFDGGFGGFGGGFFFGFF8DFDFDFFFFFFFFFBCDB<@EAFB@ABAC@CDFF?4>EEFE<*>BDAFB@FFBFF>((6<5CC.;C;=D9106(.))).)-46<<))))))))))((,(-)))()((())) NM:i:13 RG:Z:foo AS:i:82 XB:f:152.546 XE:Z:3.15e-40
(...)
- Aurélien Guy-Duché
- Pierre Lindenbaum
- Issue Tracker: https://github.com/guyduche/Blast2Bam/issues
- Source Code: https://github.com/guyduche/Blast2Bam
The project is licensed under the MIT2 license.