Physiome publishes reproducible and reusable mathematical models of physiological processes where the experimental details and model validation have been published or accepted for publication in a recognised 'primary' peer-reviewed journal. Physiome curators will help authors ensure that models and simulation experiments are made available using appropriate community standards prior to acceptance for publication in Physiome.
More details about the Physiome Journal itself can be found here:
https://journal.physiomeproject.org
https://auckland.figshare.com/articles/Presentation_COMBINE2019_pdf/9037403
The piece of software held in this repository is designed to facilitate the submission of manuscripts to the Physiome journal. Submitters are able to upload their manuscript file, specify additional metadata such as authors, funding etc. and then submit these to the journal for consideration. Subsequently, the journals curators able to use the administrative functions within the software platform to manage submissions, make editorial modifications and finally publish accepted manuscripts and models through into the Physiome figshare instance.
Videos showing briefly the functions of the software can be found on figshare here: https://figshare.com/articles/Physiome_Submission_System_-_example_videos/9868016
This development utilises and builds upon the PubSweet application core managed by the Coko Foundation and wider PubSweet community.
PubSweet Project:
https://gitlab.coko.foundation/pubsweet/pubsweet
Coko Foundation:
https://coko.foundation/
For local development the easiest way to get started is to have Docker installed and run the following services on Docker to link to the Physiome submission system instance.
- NodeJS
- Yarn
- Docker (for local development ease)
- Camunda BPM (camunda/camunda-bpm-platform:latest)
- Postgres (postgres:latest)
- Camunda Modeller (download) - for viewing/modifying BPM model documents
Create a new Postgres instance
docker run -d --name physiome-postgres -p 5432:5432 postgres:latestCreate a new Camunda BPM Engine instance
docker run -d --name physiome-camunda -p 8080:8080 camunda/camunda-bpm-platform:latestFrom the root project directory perform the following
yarn install
yarn dsl-compileThe application itself pulls configuration from several different locations:
- Within the file
/packages/app/config/default.jsan overview and the default values utilised within the application can be found. - Run-time environment can also effect the aggregated configuration. The default configuration
noted above is then supplemented with
/packages/app/config/test.jsfor example when running in the "test" environment. - Environment variables for the NodeJS application are utilised in certain defaults. These
environment variables can be provided at run-time or via the .env located
/packages/app/.env
Example ".env" file for local development:
PUBSWEET_SERVER_SECRET=<replace with JWT secret>
WORKFLOW_FILE_IDENTIFIER_DOMAIN=physiome-submission-dev.ds-innovation-experiments.com
AWS_S3_ACCESS_KEY=<replace with S3 access key>
AWS_S3_SECRET_KEY=<replace with S3 secret key>
AWS_S3_REGION=<replace with S3 region>
AWS_S3_BUCKET=<replace with S3 bucket name>
ORCID_CLIENT_ID=<replace with ORCiD API client ID>
ORCID_CLIENT_SECRET=<replace with ORCiD API client secret>
AWS_SES_ACCESS_KEY=<replace with SES access key>
AWS_SES_SECRET_KEY=<replace with SES secret key>
AWS_SES_REGION=<replace with SES region>
FIGSHARE_API_BASE=https://api.figsh.com/v2/
FIGSHARE_API_TOKEN=<replace with figshare API token>
PUBLISH_FIGSHARE_TYPE="journal contribution"
PUBLISH_FIGSHARE_GROUP_ID=<replace with figshare group id>
PUBLISH_FIGSHARE_CATEGORIES=4,12,135
DIMENSIONS_API_BASE=https://app.dimensions.ai/api
DIMENSIONS_API_USERNAME=<replace with dimensions API username>
DIMENSIONS_API_PASSWORD=<replace with dimensions API password>
STRIPE_IS_PRODUCTION=false
STRIPE_SECRET_KEY=<replace with stripe secret key>
STRIPE_PUBLISHABLE_KEY=<replace with stripe public key>
STRIPE_WEBHOOK_SECRET_KEY=<replace with stripe web hook secret key>
IDENTITY_ADMIN_USERS=<comma separated list of ORCID IDs for admin users>
EMAIL_SEND_FROM=Physiome Journal <physiome-curators@physiomeproject.org>
EMAIL_SUBJECT_PREFIX="Physiome submission system:"
EMAIL_SIGNATURE="Kind regards,\nPhysiome curation team\nphysiome-curators@physiomeproject.org"
EMAIL_RESTRICTED_TO=<replace with restricted email addresses for dev/test environment, empty for production>
EMAIL_EDITORS_MAILING_LIST="physiome-editors@physiomeproject.org"
- The business logic is described within
/definitions/physiome-submission-v2.bpmn. This file can be opened using the Camunda Modeler application. It describes the tasks and steps a submission goes through, including any "external tasks" such as sending an email or publishing to figshare. - The description of how these components all fit together (the glue so to speak) is defined within the file
/definitions/physiome-submission.wfd. This contains a custom domain-specific language that is "compiled" via PegJS into a JSON file representation. The compiled representation can be found within/packages/app/config/description.json
The minimum required to get this configured locally for testing things out is to create the .env file and provide it with the required values for S3, SES and ORCID. The figshare options do not need to be specified for local development purposes (unless you have access to a test figshare instance).
For deployment purposes or to utilise a database/BPM from another machine, it is also possible to define a Postgres database and Camunda engine located elsewhere using the following environment variables:
DATABASE=<postgres database name>
DB_USER=<postgres username>
DB_PASS=<postgres password>
DB_HOST=<postgres server hostname>
WORKFLOW_API_URI=http://<camunda hostname>:8080/engine-rest
We need to compile the current "Workflow Description". This will use the PegJS created processor to
read in the file /definitions/award-submission.wfd and process it into the description JSON
file located within /packages/app/config/description.json
yarn descIn order for Camunda to know about the business logic model we wish to use, it needs to be deployed to the Camunda instance. This will happen automatically as part of a Knex database migration script.
With all of the above configured and deployed it is now possible to run the application.
From the root directory of the project, run the following command:
yarn startThis will kick off the initial database setup steps (including BPM deployments) and then run the backend GraphQL endpoint. It will also run webpack over the front-end application and expose it for testing.
Navigate in your local browser of preference to: http://localhost:3000
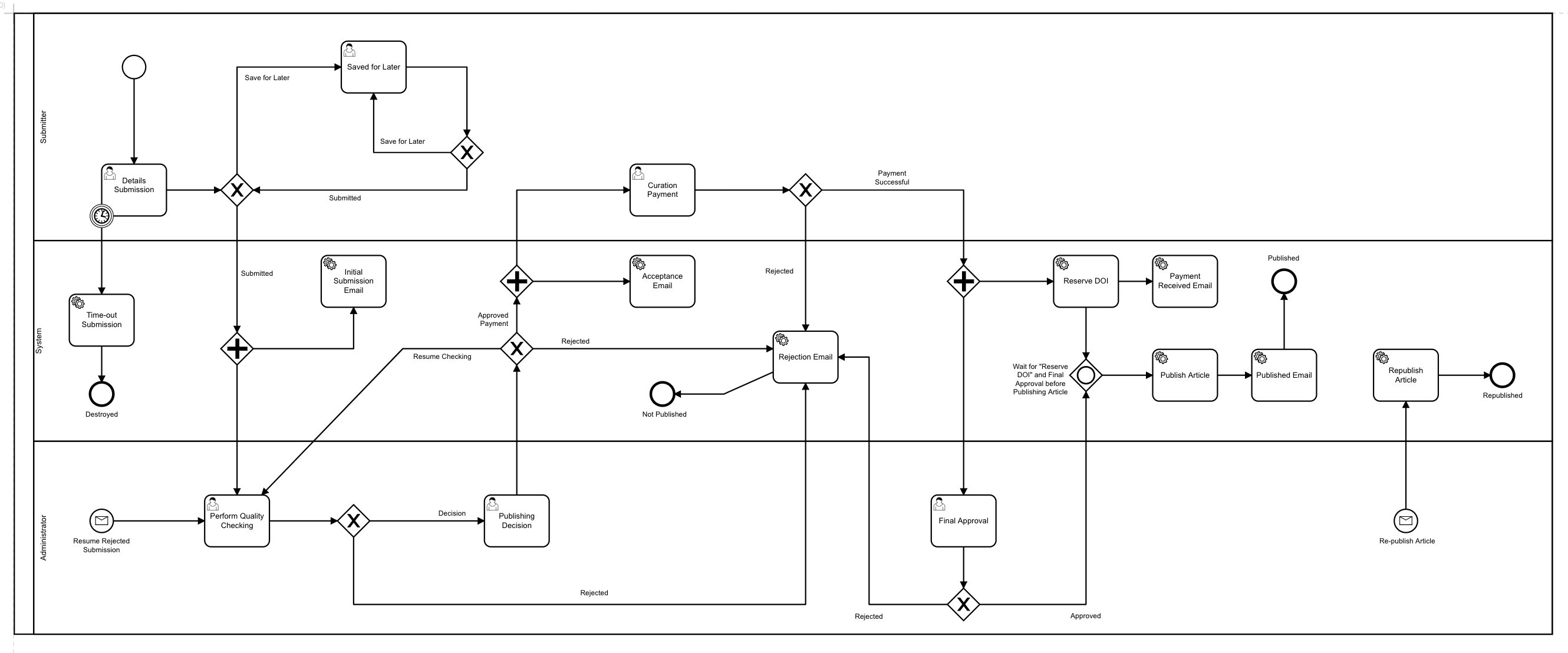
The BPMN workflow (as seen above from the Camunda Modeller) describes the workflow and possible actions at each stage a submission may go through during its lifetime. It also describes any third-party integrations, defining where and when they are enacted during the process of a submission. Each task within the BPM definition has specific attributes which tie into the workflow description (expressed in our custom DSL) which defines what form is associated with a task. The workflow description also includes details relating to user interface elements, data models, validations and access control restrictions on data.
The above example workflow description shows validations that are applied during the manuscript submission phase, including the end-user warnings that are displayed when a validation fails. The example layout also shows how the submission form itself is generated, including the links between a form element and the data model it targets.
The code relating to how the DSL language takes the data model and transforms this into a representation used within the PubSweet core is in need of a refactoring, so please ignore the code smell that lingers in these parts.
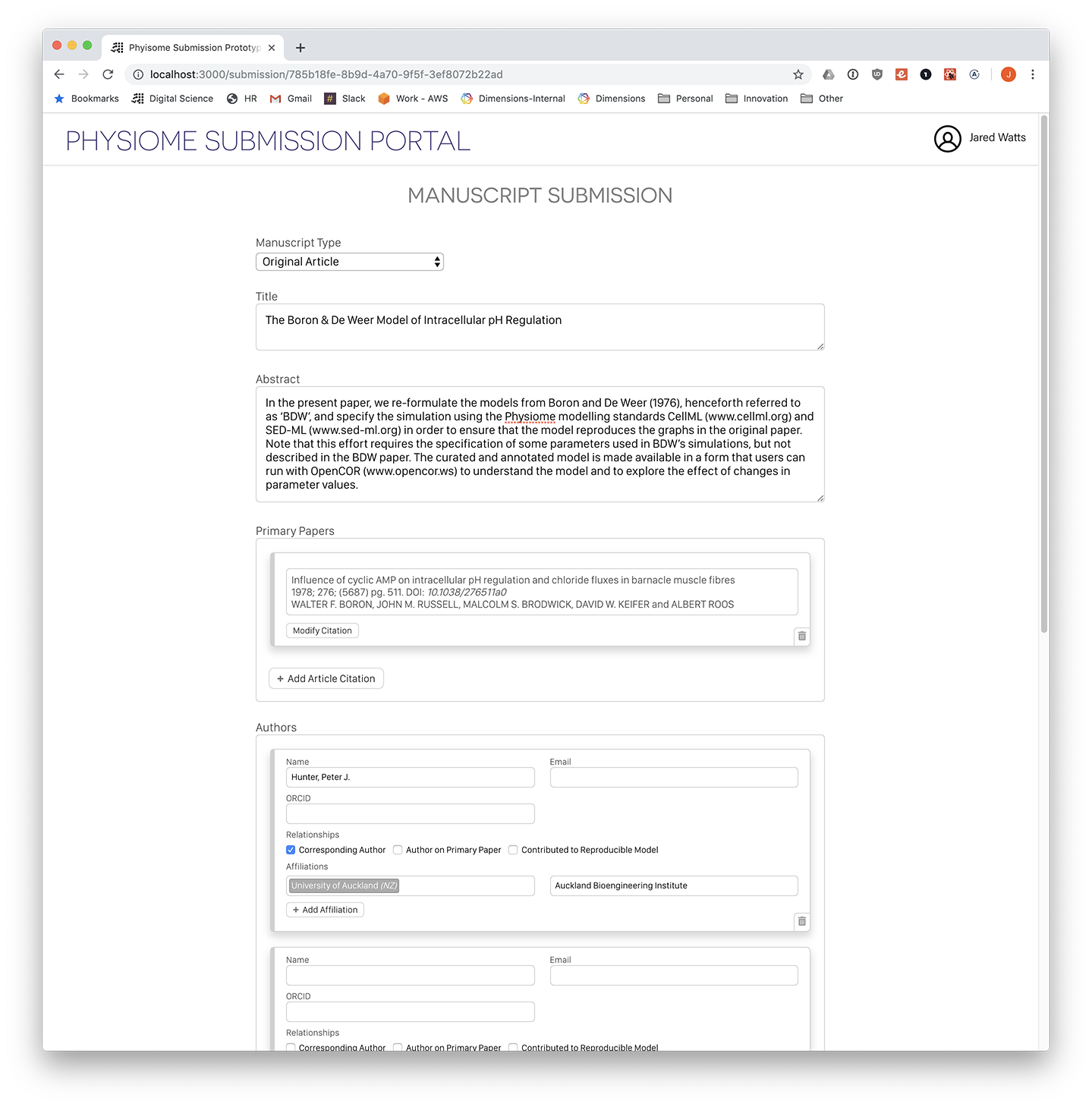
Screenshot: Manuscript submission form
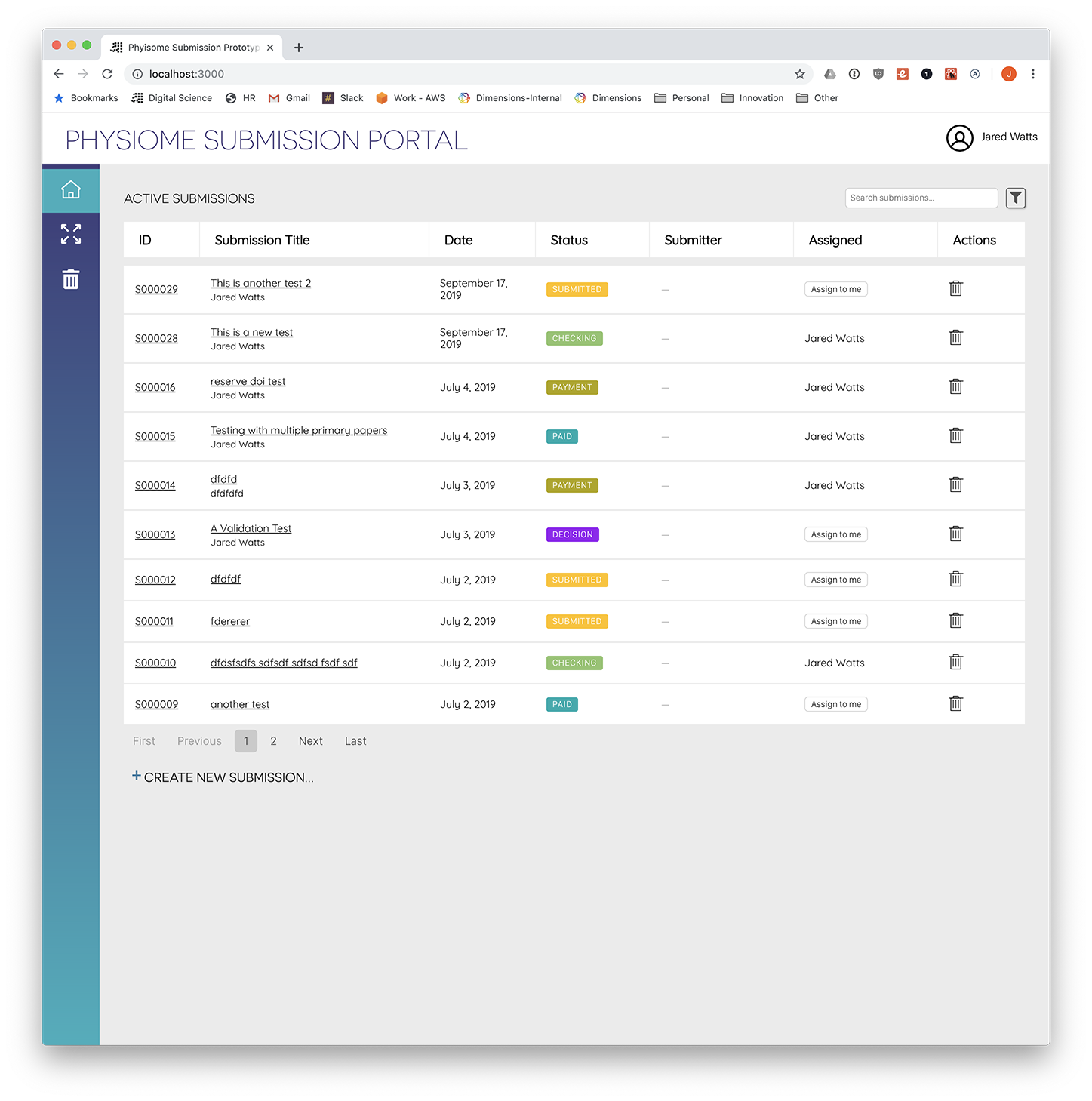
Screenshot: Administration view with submissions listing
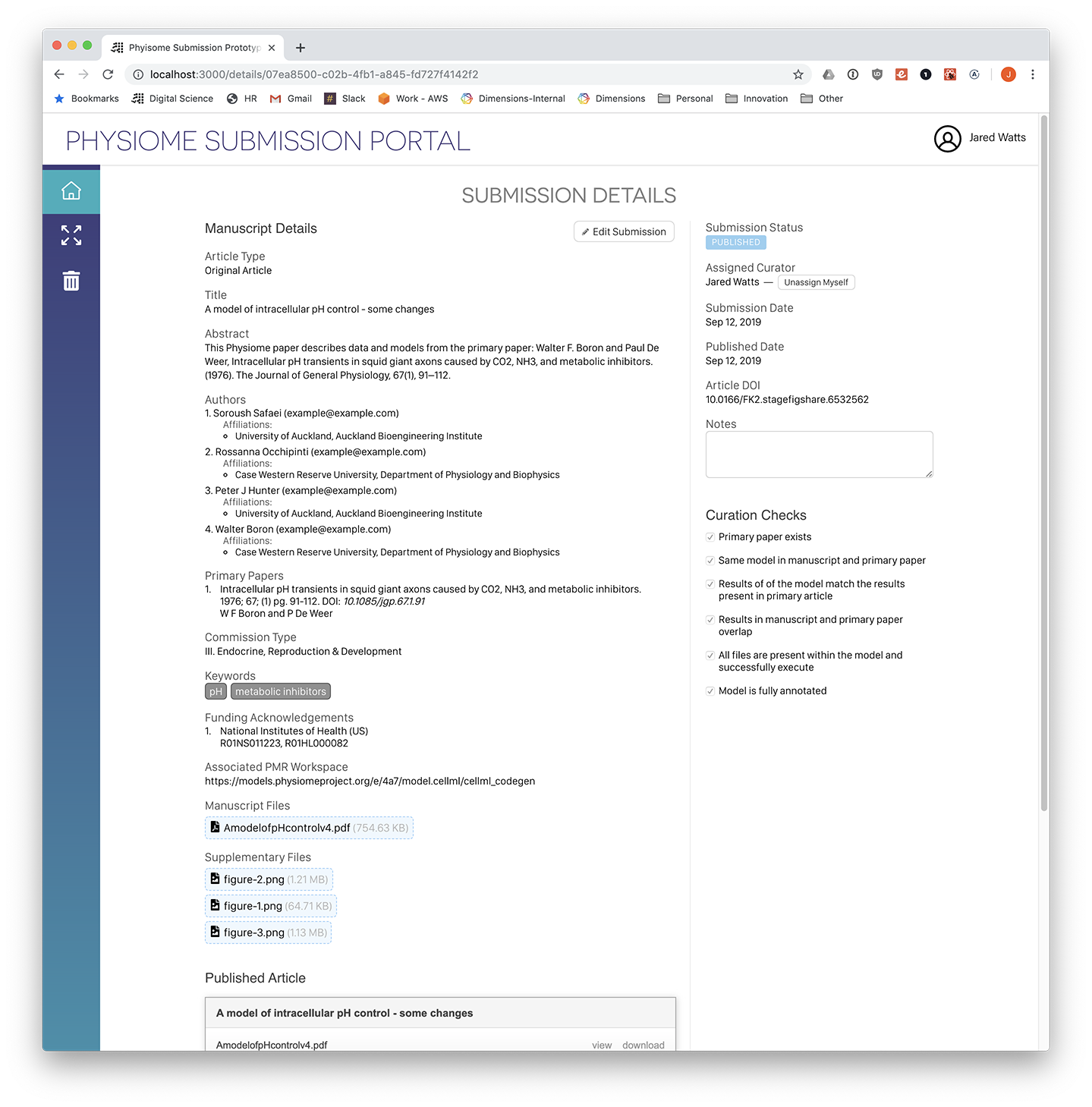
Screenshot: Administration view, submission details
Videos of functionality can be seen here: https://figshare.com/articles/Physiome_Submission_System_-_example_videos/9868016
Please feel free to contact Jared Watts (j.watts@digital-science.com) about this project. To contact the Physiome journal, physiome@physiomeproject.org.