-
Write a function that returns a float corresponding to the volume of a sphere:
get_sphere_volume(radius) (formula=(4/3)πr 3 ) -
Write a function that calculates and returns an integer corresponding to the factorial of an integer (n):
a) Using recursivity: recursive_factorial(n) b) Without using recursivity: factorial(n) -
Write a function for counting up numbers from 0 to n, showing the count up in the screen. If parameter odd is set to True, prints only odd numbers
a) Using recursivity: recursive_count_up(n, odd) b) Without using recursivity: count_up(n,odd) -
Find and solve the bugs in the following function:
def get_final_price(discount_percentage=10, price): """Return the final price after applying the discount percentage """ return ((price + price) * percentage) / 100
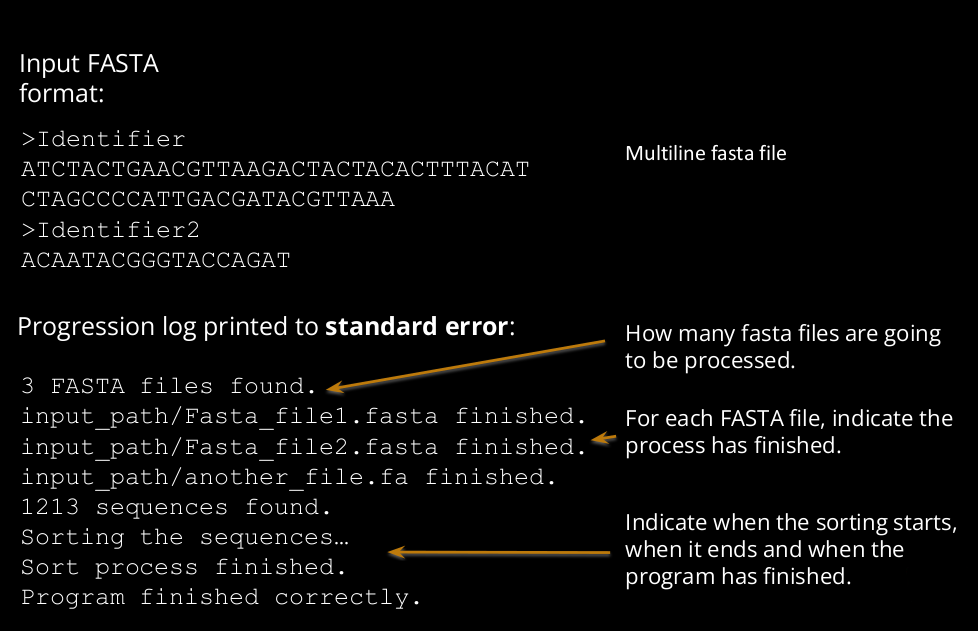
Input is a multiline FASTA file.
-
Given a multi-line protein FASTA file (stored in a file with path defined filename), returns a float corresponding to the ratio of proteins in the fasta file having a relative frequency higher or equal than a given threshold provided as an argument named “relative_threshold” and having an absolute frequency of the same residue higher or equal than a given threshold provided as an argument named “absolute_threshold” for a given residue. The function should be named as follows, with the same arguments definition:
get_proteins_ratio_by_residue_threshold(filename, residue,relative_threshold=0.03, absolute_threshold=10) -
Given a protein FASTA file (filename), save on a output file named output_filename the protein identifier, the first N-aminoacids, the last M-aminoacids and the absolute frequency in the protein of all the aminoacids found in the protein (the aminoacids that do not appear in the protein should not be shown). The fields must be separated by a tabulator, and one protein by line.
print_sequence_summary(filename, output_filename, first_n=10, last_m=10)
Example:
Input:
>PROT1
EFTRPTSTWSAAALMTRSSSTRWSPD
>PROT2
SSTPLRRSTPAWEEFGLMCCDPRS
>PROT3
ATRSLEWKSTPW
Output:
PROT1 EFT RWSPD E:1,F:1,T:5,R:3,P:2,S:6,W:2,A:3,L:1,M:1,D:1
PROT2 SST CDPRS S:4,T:2,P:3,L:2,R:3,A:1,W:1,E:2,F:1,G:1,M:1,C:2,D:1
PROT3 ATR KSTPW A:1,T:2,R:1,S:2,L:1,E:1,W:2,K:1,P:1
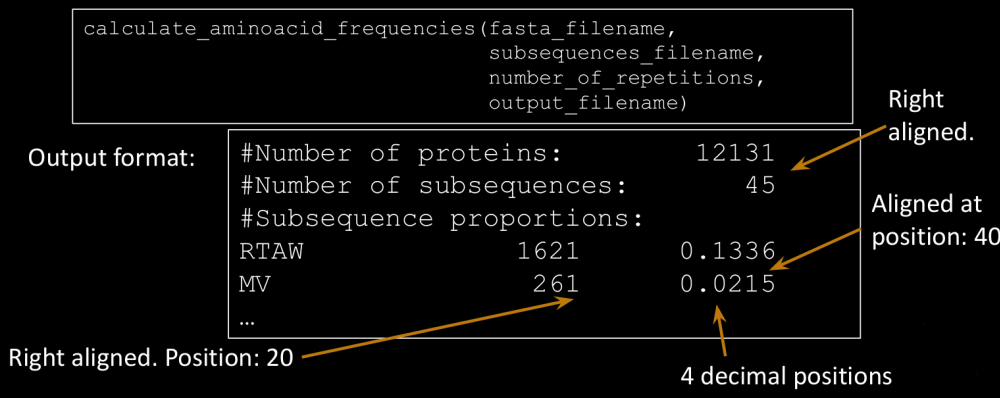
- Create a function that, given a multi-line protein FASTA file (fasta_filename) and a “sub-sequences” file (subsequences_filename) (one sequence in each line), calculates the proportion of proteins in the FASTA file containing at least N-times (number_of_repetitions) each of the sub-sequences (exactly equal). Save it in an output file with the specified format, ordered by the proportion value (descending order)
-
A Generator Function that reads a Fasta file. In each iteration, the function must return a tuple with the following format: (identifier, sequence). Function name:
FASTA_iterator(fasta_filename) -
Given a list of FASTA files, create a function that returns a dictionary that contains the 4 following keys with the associated values:
- “intersection”: a set with the common identifiers found in all the files
- “union”: a set with all the identifiers (unique) found in all the files
- “frequency”: a dictionary with all the identifiers as keys and the number of files in which it appears as values (int)
- “specific”: a dictionary with the name of the input files as keys and a set with the specific identifiers as values (i.e. identifiers that are exclusive in that fasta file)
Note 1: Common identifier equivalence must be case-insensitive (i.e. Code_A,code_a and
CODE_A are equivalents).
Note 2: It must use the FASTA_iterator function created in exercise 1.
Function name:
compare_fasta_file_identifiers( fasta_filenames_list )
Note: Use the FASTA_iterator function created in last exercises.
-
Repeat the same exercises proposed in session 2 but using the FASTA_Iterator function created in session 4 to read the FASTA files.
-
A function that, given a multiline FASTA file, returns the length of the sequence with the maximum length
get_max_sequence_length_from_FASTA_file( fasta_filename ) -
A function that, given a multiline FASTA file, returns the length of the sequence with the minimum length
get_min_sequence_length_from_FASTA_file ( fasta_filename ) -
A function that, given a FASTA file, returns a list of tuples (identifier, sequence) corresponding to the sequence(s) with maximum length. The list must be sorted by the identifier (case insensitive sorted).
get_longest_sequences_from_FASTA_file( fasta_filename ) -
A function that, given a FASTA file, returns a list of tuples (identifier, sequence) corresponding to the sequence(s) with minimum length. The list must be sorted by the identifier (case insensitive sorted).
get_shortest_sequences_from_FASTA_file( fasta_filename ) -
A function that, given a protein FASTA file, returns a dictionary with the molecular weights of all the proteins in the file. The dictionary keys must be the protein identifiers and the associated values must be a float corresponding to the molecular weight.
get_molecular_weights( fasta_filename ) -
A function that, given a protein FASTA file, returns a tuple with (identifier, sequence) of the protein with the lowest molecular weight. If there are two or more proteins having the minimum molecular weight, just return the first one.
get_sequence_with_min_molecular_weight( fasta_filename ) -
A function that, given a protein FASTA file, returns the mean of the molecular weights of all the proteins
get_mean_molecular_weight( fasta_filename )
Create a python function that calculates the mean of the minimum distance between any two residues pairs found in the same chain of a PDB. The script, when executed by command line, should output in standard output the mean distance for each chain (with 4 decimal positions). The python script should use a single argument corresponding to the PDB file path to use. This command line argument is optional. If the PDB file path is not defined, read the PDB file from standard input. The function must return a dictionary with chains as keys and mean minimum distances as values. It uses a single argument which specifies the path of the PDB file If the argument pdb_file_path is None, read the PDB file from standard input.
calculate_pdb_chain_mean_minimum_distances(pdb_file_path)
When the file is imported as a module, it should not execute the function. The function should only be called when the script is executed by command line.
Sample output in command line:
A: 22.7400
B: 20.4224
N: 23.9393
F: 23.9730
J: 23.4187
-
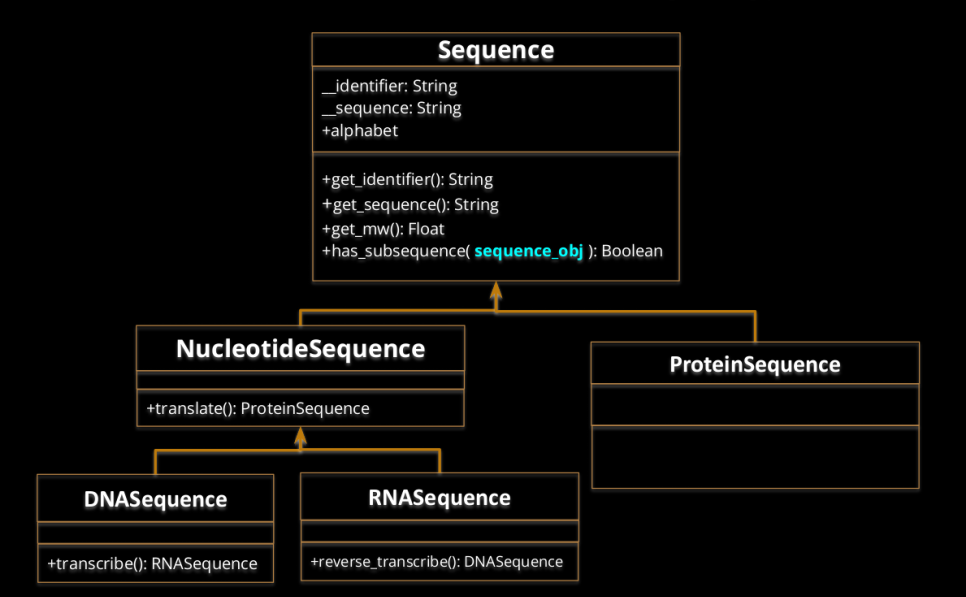
Define a new class named Protein, with the following definition. If necessary, you can define the private methods you need
# Protein +identifier: String +sequence: String --- +get_identifier(): string +get_sequence(): string +get_mw(): float +has_subsequence( Protein): boolean +get_length(): integer -
Modify the FASTA_iterator generator function to yield Protein objects instead of tuples. In each iteration, the function must yield a Protein Object: FASTA_iterator( fasta_filename )
- Define and implement the following classes:
Specifications:
- alphabet must be a class attribute that specifies the possible alphabet of the sequence.
- When creating a new Sequence instance (ProteinSequence, DNASequence or RNASequence), it must check that the sequence is correct by checking in the alphabet. If not, raise an exception with the following statement, where X is the incorrect letter: raise ValueError(“Impossible to create instance: X not possible”)
- If necessary, additional attributes or methods can be created, private or public.
- You can find required data in a file called sequence_dictionaries.py
Define the following behaviour:
- len(Sequence): should return the length of the sequence.
- sequence1 == sequence2: return True if sequence strings are exactly the same (without taking into account the identifiers).
- sequence1 != sequence2: return True if sequences are different, without taking into account the identifiers.
- Sequence + Sequence: Create a new sequence object instance with their sequences concatenated. Sequence object has to be of the same class as the operands. It should not be applicable to different classes (i.e. ProteinSequence, RNASequence). The identifiers should also be concatenated with a “+” as a glue between both identifiers.
- Sequence[i]: should return the sequence element at position i. Position 0 corresponds to the first position.
- in operator: should return a boolean if the string is a substring of the attribute sequence.
- Comparing sequences. Implement the necessary method(s) to define how sequences should be ordered. The objective is that when sorting a list of sequences, they are sorted according to their molecular weight.
- Adapt the sequence class so that it can be used as key in a dictionary or it can be added to a set. Two sequences should be considered the same object in terms of set or key if they share both the identifier and the sequence.
-
Create a new ValueError exception subclass named IncorrectSequenceLetter
-
To create a new exception instance, it should be created with the letter not found in the alphabet and the class name of the sequence. Example: e = IncorrectSequenceLetter(“B”, class_name)
-
The description string of the exception must be the following: “The sequence item B is not found in the alphabet of class ProteinSequence”
-
Sequence class should raise an IncorrectSequenceLetter exception when a sequence is created using an incorrect letter not found in the alphabet.
-
Modify the FASTA_iterator to be able to iterate in any type of Sequence (add a new argument in the generator function to specify the Sequence class to use) ) Modify the FASTA_iterator generator function to skip sequences having incorrect letters . It must capture specifically the IncorrectSequenceLetter exception. When it happens, it should print a message of error in the standard error and continue with the next sequence. Do not handle other types of exceptions, only IncorrectSequenceLetter exceptions.
-
When the script is executed as a standalone application (without being imported (i.e. code under main block), the script should read input DNA FASTA file(s) and calculate the length and molecular weight of their corresponding proteins (i.e. corresponding to ProteinSequence instances obtained after translation). The script must print the output to standard output or to a file. Output should be sorted by molecular weight, from lowest to greatest.
python3 script.py [IN] [OUT]
7.1) If the script is executed without arguments, it looks for all “.fasta” or “.fa” files in the current directory, and process all of them with a single sorted output. Print the results to standard output.
7.2) If the script has a single argument, it corresponds to the input. If it is a directory, it looks for all “.fasta” or “.fa” files in the given directory, process them and print the results in standard output. If this single argument corresponds to a file (not necessarily “.fasta”, or “.fa”), process it and print the results in standard output.
7.3) If the script has two arguments, the first one corresponds to the input and the second one to the output file.
7.4) The script should print to standard error a progression log
Output format:
Identifier1 Length1 molecular_weight1
Identifier2 Length2 molecular_weight2
Identifier3 Length3 molecular_weight3