- PyTorch 1.1
- TorchProteinLibrary (https://github.com/lupoglaz/TorchProteinLibrary, commit 16166ce4847ad3ba95cd5afdb8bb4503cad32caa)
- SE3CNN (https://github.com/mariogeiger/se3cnn)
The directories are defined in src/init.py.
The instruction on how to prepare the dataset can be found here
-
Dataset loaders can be found in src/Dataset directory. We use two loaders: one for the generated dataset and one for the DockingBenchmark.
-
Models are defined in src/Models. They are split into Protein Representation Models and Docking Models. Additionally, here one can find the ranking loss function and volume multiplication modules
-
Training iteration is defined in src/Training/LocalTrainer.py
-
Docking iteration is defined in src/Docker/Docker.py
Main scripts for training and testing are: src/train_local.py and src/test_local.py. Training and testing parameters we used in the paper can be found in src/server_train_local.sh and src/server_test_local.sh
- Rotations loading and circular volume plotting can be found in src/Utils
We used rotation generated by https://mitchell-lab.biochem.wisc.edu/SOI/index.php that can be found in data directory. However, they have different license than this repository. Be sure to obtain the license from MitchellLab.
To plot training curve one has to run script scripts/Results/PlotTraining.py. The generated loss curve example:
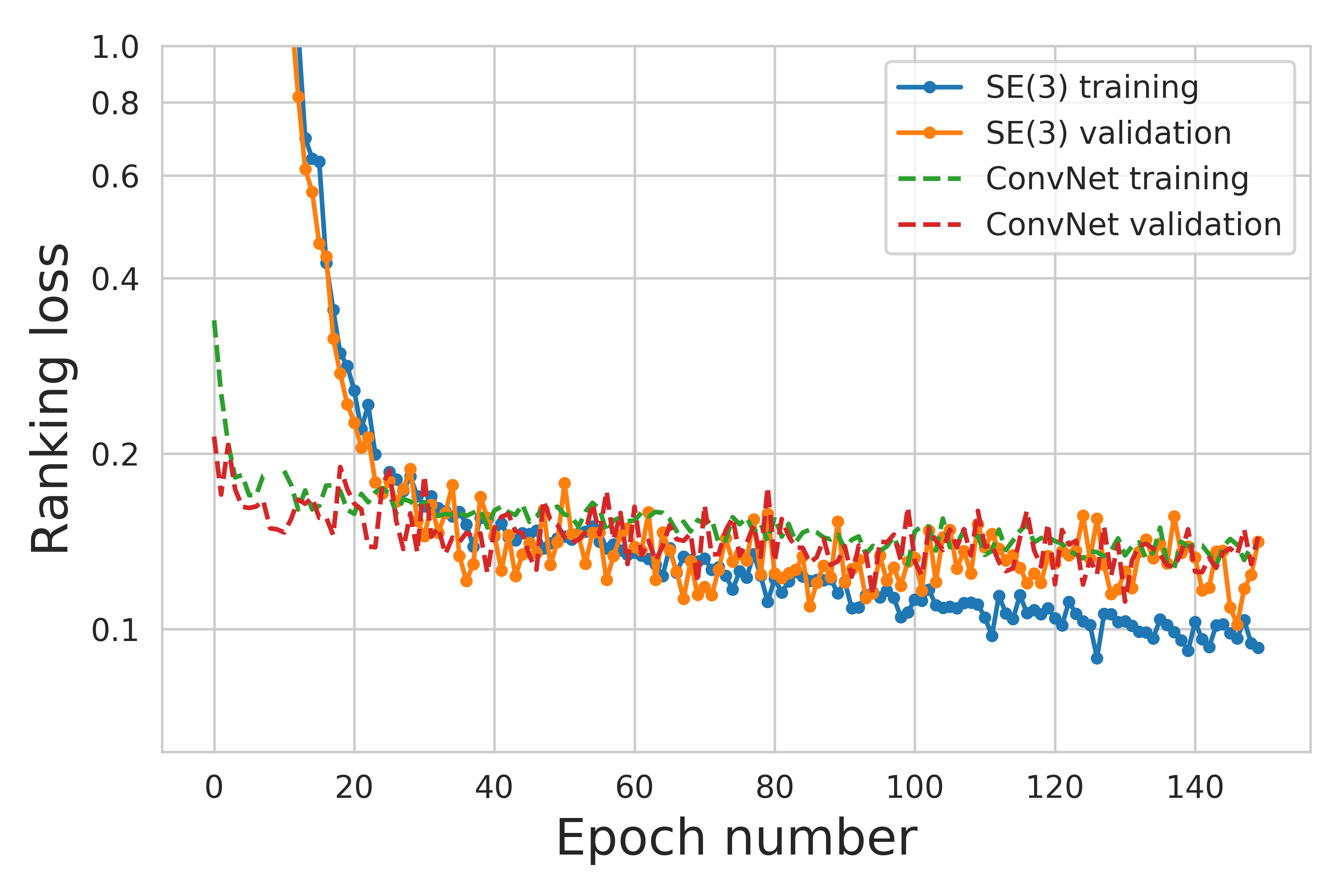
The benchmark results can be computed using script scripts/Results/PlotBenchmark.py. Refactoring, work in progress.