The interactive learning and segmentation toolkit
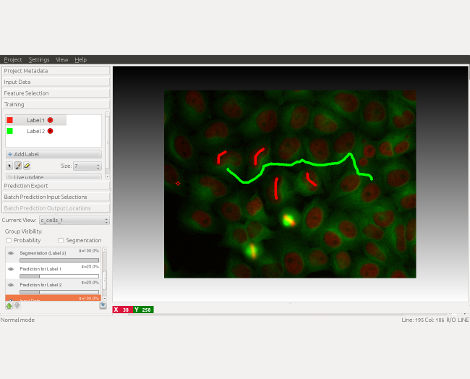
Leverage machine learning algorithms to easily segment, classify, track and count your cells or other experimental data. Most operations are interactive, even on large datasets: you just draw the labels and immediately see the result. No machine learning expertise required.
See ilastik.org for more info.
Go to the download page, get the latest non-beta version for your operating system, and follow the installation instructions. If you are new to ilastik, we suggest to start from the pixel classification workflow. If you don't have a dataset to work with, download one of the example projects to get started.
ilastik is also available as a conda package on our ilastik-forge conda channel.
We recommend using mamba instead of conda, for faster installation:
mamba create -n ilastik --override-channels -c pytorch -c ilastik-forge -c conda-forge ilastik
# activate ilastik environment and start ilastik
conda activate ilastik
ilastikVersions of ilastik until 1.4.1b2 are based on, and only compatible with Python 3.7
Starting from ilastik 1.4.1b3 ilastik environments can be created with Python versions 3.7 to 3.9.
Limitations when going with Python 3.7: please use a version of tifffile >2020.9.22,<=2021.11.2 (see also note in environment-dev.yml).
ilastik is a collection of workflows, designed to guide you through a sequence of steps. You can select a new workflow, or load an existing one, via the startup screen. The specific steps vary between workflows, but there are some common elements like data selection and data navigation. See more details on the documentation page.
If you have a question, please create a topic on the image.sc forum. Before doing that, search for similar topics first: maybe your issue has been already solved! You can also open an issue here on GitHub if you have a technical bug report and/or feature suggestion.
We always welcome good pull requests! If you just want to suggest a documentation edit, you can do this directly here, on GitHub. For more complex changes, see CONTRIBUTING.md for details.