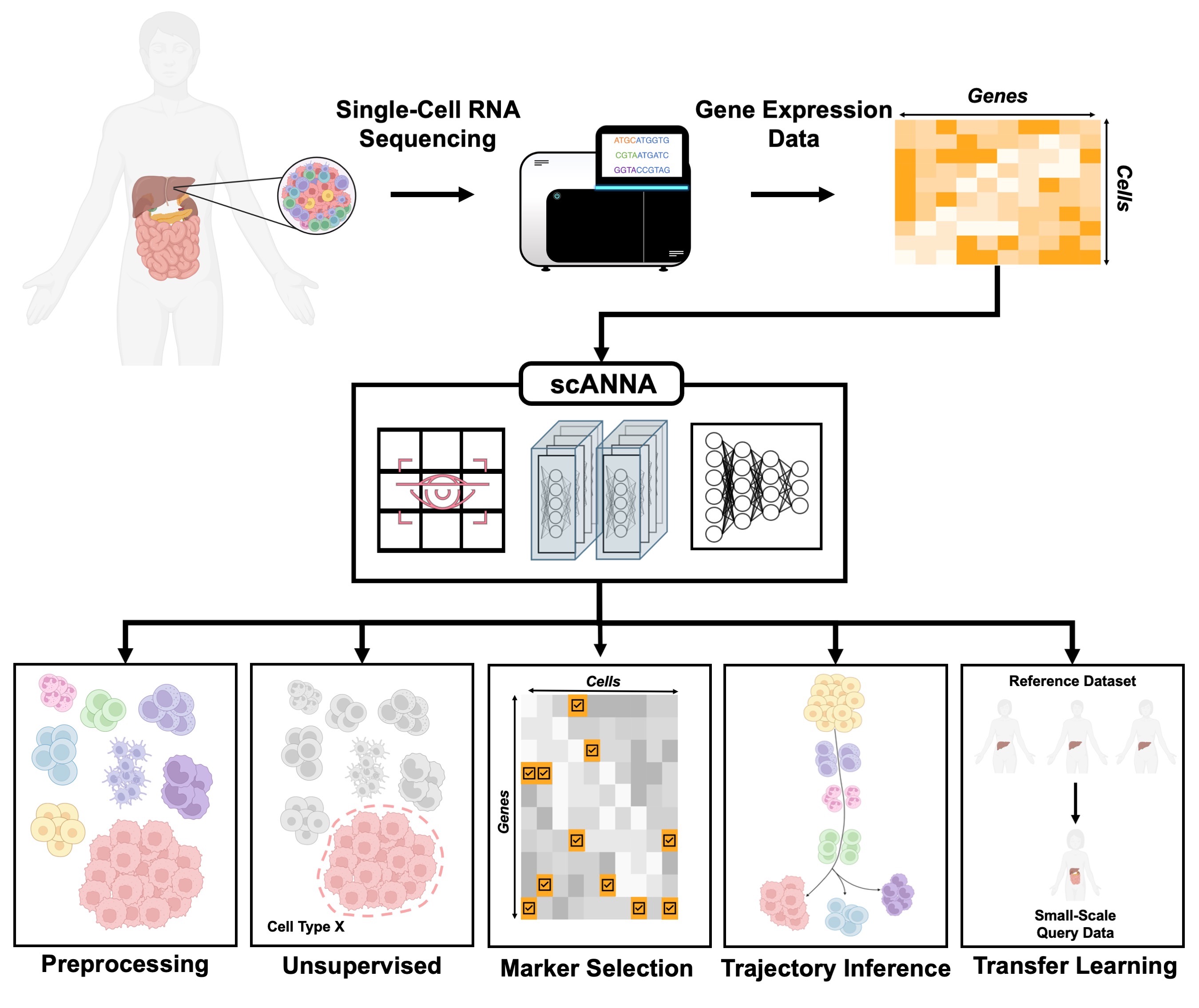
This repository hosts the package for scANNA: single-cell ANalysis using Neural Attention. To make package maintenance more efficient, and to provide more specific tutorials on using scANNA, we have located tutorials in a dedicated repository, as listed below.
scANNA requires Python 3.10 and can be installed using PyPI:
$ pip install git+https://github.com/SindiLab/scANNA.git
or can be first cloned and then installed as the following:
$ git clone https://github.com/SindiLab/scANNA.git
$ pip install ./scANNA
Once the files are available, make sure to be in the same directory as setup.py. Then, using pip, run:
pip install -e .In the case that you want to install the requirements explicitly, you can do so by:
pip install -r requirements.txtAlthough the core requirements are listed directly in setup.py. Nonetheless, it is good to run this beforehand in case of any dependecies conflicts.
All main scripts for training (and finetuning) our deep learning model are located in the training_and_finetuning_scripts folder in this repository.
We have compiled a set of notebooks as tutorials to showcase scANNA's capabilities and interptretability. These notebooks located here.
Please feel free to open issues for any questions or requests for additional tutorials!
TODO: Will be released with the next preprint for scANNA.
If you found our work useful for your ressearch, please cite our preprint:
@article {Davalos2023.05.29.542760,
author = {Oscar A. Davalos and A. Ali Heydari and Elana J Fertig and Suzanne Sindi and Katrina K Hoyer},
title = {Boosting Single-Cell RNA Sequencing Analysis with Simple Neural Attention},
elocation-id = {2023.05.29.542760},
year = {2023},
doi = {10.1101/2023.05.29.542760},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2023/06/01/2023.05.29.542760},
eprint = {https://www.biorxiv.org/content/early/2023/06/01/2023.05.29.542760.full.pdf},
journal = {bioRxiv}
}