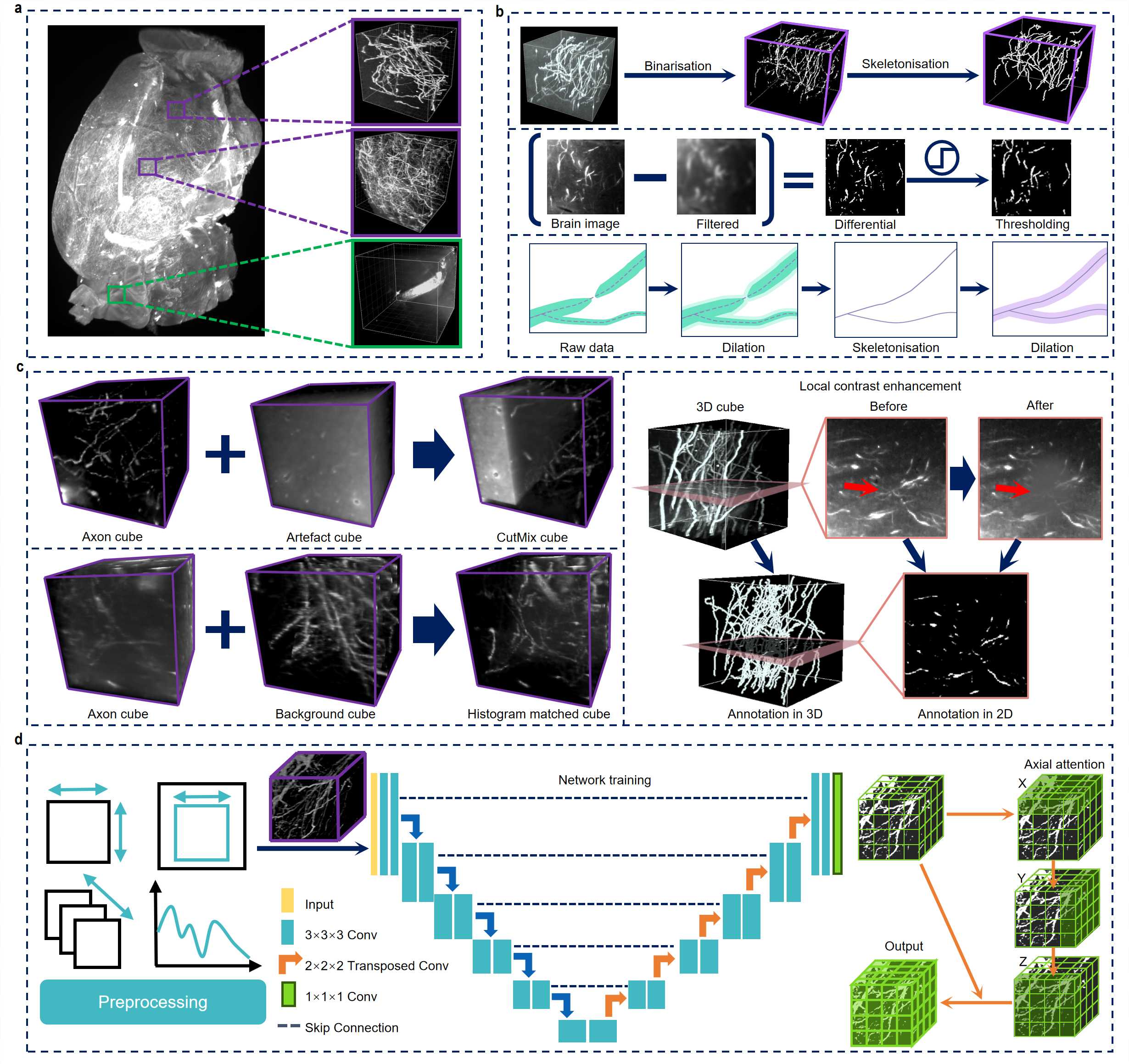
This is the repo for the whole-brain axon segmentation of the paper D-LMBmap: a fully automated deep-learning pipeline for whole-brain profiling of neural circuitry
- An end-to-end pipeline for whole-brain axon segmentation and profiling
- Five types of neurons, including Serotonergic, GABAergic, Dopaminergic, and Cerebellar nuclei neurons, are used to train the automated segmentation models separately and jointly
- An automated annotation toolkit is provided, greatly reducing the labour-intensive manual input
Recent proliferation and integration of tissue-clearing methods and light-sheet fluorescence microscopy has created new opportunities to achieve mesoscale three-dimensional whole-brain connectivity mapping with exceptionally high throughput. With the rapid generation of large, high-quality imaging datasets, downstream analysis is becoming the major technical bottleneck for mesoscale connectomics. Current computational solutions are labor intensive with limited applications because of the exhaustive manual annotation and heavily customized training. In this repository, we provide an automated pipeline for axon data annotation and segmentation, greatly reducing the workload of labour-intensive manual input. In addition, we offer segmentation models for five distinct types of axons, as well as an jointly trained axon segmentation model with enhanced generalizability. Researchers can use these models for fine-tuning in subsequent personalized tasks.
The data volume information for the five datasets is as follows. We have trained segmentation models for each type of axon using these five datasets. Additionally, we have also trained a more generalized comprehensive model using a combined dataset from all five categories (4714 cubes after data augmentation). Please download these models via the provided model link based on your specific needs.
| Neuron type | Whole-brain resolution | No. of axon cubes | No. of artefact cubes | No. of cubes after data augmentation | Cube Size | Data link |
|---|---|---|---|---|---|---|
| Serotonergic sparse stained | 46 | 54 | 1040 | link | ||
| Cerebellar nuclei sparse stained | 49 | 47 | 725 | link | ||
| Serotonergic dense stained | 86 | 10 | 1024 | link | ||
| GABAergic dense stained | 91 | 45 | 1452 | link | ||
| Dopaminergic dense stained | 84 | 100 | 1156 | link |
torch==1.11.0
torchvision==0.12.0
nnUNet==1.7.0
timm==0.9.7
git clone git@github.com:lmbneuron/D-LMBmap.git
cd "Axon Segmentation"
cd "Axon segmentation model training"
pip install -e .
pip install -r requirements.txtOur models are built based on nnUNet. Apart from above installation, please ensure that you meet the requirements of nnUNet.
Please download pre-trained models via the provided model link based on your specific needs.
Before your training, make sure you have prepared the training cubes with automatically annotated masks and stored them as below.
Run create_data.py, in which base and source directories should be prepared ahead as below. Note that the number of
the skeletonized annotations, the automatically annotated masks and the axon cubes should be exactly the same.
└── base(original training data)
└── train
├── cropped-cubes(training axon cubes)
│ └──volume-001.tiff
├── Rough-label(automatically annotated masks)
│ └──label-001.tiff
├── Fine-label(skeletonized annotation)
│ └──label-001.tiff
└── artifacts(junk cubes)
└──volume-200.tiff
└── source(data used for histogram matching)
└── train
├── cropped-cubes
│ └──volume-001.tiff
├── Rough-label
│ └──label-001.tiff
├── Fine-label
│ └──label-001.tiff
└── artifacts
└──volume-200.tiff
We propose three data augmentation methods, histogram matching, cutmix, and local contrast enhancement to augment training data.
Change the parameters of function histogram_match_data in create_data.py to choose using histogram matching/cutmix/contrast enhancement or
not. If you want to use histogram matching, it is better to set both match_flag and join_flag True so that both
original cubes and matched cubes can be used for training.
cutmix=True # use cutmix, mix up axon cubes and artifact cubes
match_flag=True, join_flag=True # use histogram matching, join matched and original cubes
match_flag=True, join_flag=False # use histogram matching, use only matched cubes
To run create_data.py:
python create_data.py --base BASE_PATH --source SOURCE_PATH --task_id ID --task_name TaskXXX_MYTASK
After last step the raw training dataset will be in the folder prepared in DATASET/raw_data_base/nnUNet_raw_data/TaskXXX_MYTASK,
where task id XXX and task name MYTASK are set in create_data.py.
For training our model, a preprocessing procedure is needed. Run this command:
nnUNet_plan_and_preprocess -t XXXYou will find the output in DATASET/preprocessed/TaskXXX_MYTASK.
There are several additional input arguments for this command. Running -h will list all of them along with a description. If you run out of RAM during preprocessing, you may want to adapt the number of processes used with the -tl and -tf options. The default configuration make use of a GPU with 8 GB memory. Larger memory size can be used with options like -pl3d ExperimentPlanner3D_v21_16GB.
Our model trains all U-Net configurations in a 5-fold cross-validation. This enables the model to determine the postprocessing and ensembling on the training dataset.
Training models is done with the nnUNet_train command. The general structure of the command is:
nnUNet_train CONFIGURATION TRAINER_CLASS_NAME TASK_NAME_OR_ID FOLD --npz (additional options)CONFIGURATION is a string that identifies the requested U-Net configuration. TASK_NAME_OR_ID specifies what dataset should be trained on and FOLD specifies which fold of the 5-fold-cross-validaton is trained.
TRAINER_CLASS_NAME is the name of the model trainer. To be specific, a normal U-Net will be trained with TRAINER_CLASS_NAME nnUNetTrainerV2.
We also propose networks with attention modules. You can use TRAINER_CLASS_NAME MyTrainerAxial to train a U-Net with attention modules. If you need to continue a previous training, just add a -pretrained_weights to the training command. For FOLD in [0, 1, 2, 3, 4], a sample command is:
nnUNet_train 3d_fullres MyTrainerAxial TaskXXX_MYTASK FOLD -p nnUNetPlansv2.1_16GB --pretrained_weights PRETRAINED_MODEL
The trained models will be written to the DATASET/trained_models/nnUNet folder. Each training obtains an automatically generated output folder name DATASET/preprocessed/CONFIGURATION/TaskXXX_MYTASKNAME/TRAINER_CLASS_NAME__PLANS_FILE_NAME/FOLD. Multi GPU training is not supported.
Once all 5-fold models are trained, use the following command to automatically determine what U-Net configuration(s) to use for test set prediction:
nnUNet_find_best_configuration -m 3d_fullres -t XXX --strictThis command will print a string to the terminal with the inference commands you need to use. The easiest way to run inference is to simply use these commands. For each of the desired configurations(e.g. 3d_fullres), run:
nnUNet_predict -i INPUT_FOLDER -o OUTPUT_FOLDER -t TASK_NAME_OR_ID -m CONFIGURATION --save_npz
Only specify --save_npz if you intend to use ensembling. --save_npz will make the command save the softmax probabilities alongside of te predicted segmentation masks requiring a lot of disk space. You can also use -f to specify folder id(s) if not all 5-folds has been trained. --tr option can be used to specify TRAINER_CLASS_NAME, which should be consistent with the class used in model training. A sample command using an U-Net with attention module to generate predictions is:
nnUNet_predict -i INPUT_FOLDER -o OUTPUT_FOLDER -t XXX --tr MyTrainerAxial -m 3d_fullres -p nnUNetPlansv2.1_16GB
We extract the model weights from the saved checkpoint files(e.g. model_final_checkpoint.model) to pth files by running python save_models.py. The pth file will be used for whole brain axon prediction.
If you encounter any issues, please contact us at lipeiqi@stu.xjtu.edu.cn
This work is licensed under a Creative Commons Attribution 4.0 International License
Our code is based on the nnU-Net framework.
If you find this repository useful, please consider citing this paper:
@article{li2023d,
title={D-LMBmap: a fully automated deep-learning pipeline for whole-brain profiling of neural circuitry},
author={Li, Zhongyu and Shang, Zengyi and Liu, Jingyi and Zhen, Haotian and Zhu, Entao and Zhong, Shilin and Sturgess, Robyn N and Zhou, Yitian and Hu, Xuemeng and Zhao, Xingyue and others},
journal={Nature Methods},
pages={1--12},
year={2023},
publisher={Nature Publishing Group US New York}
}