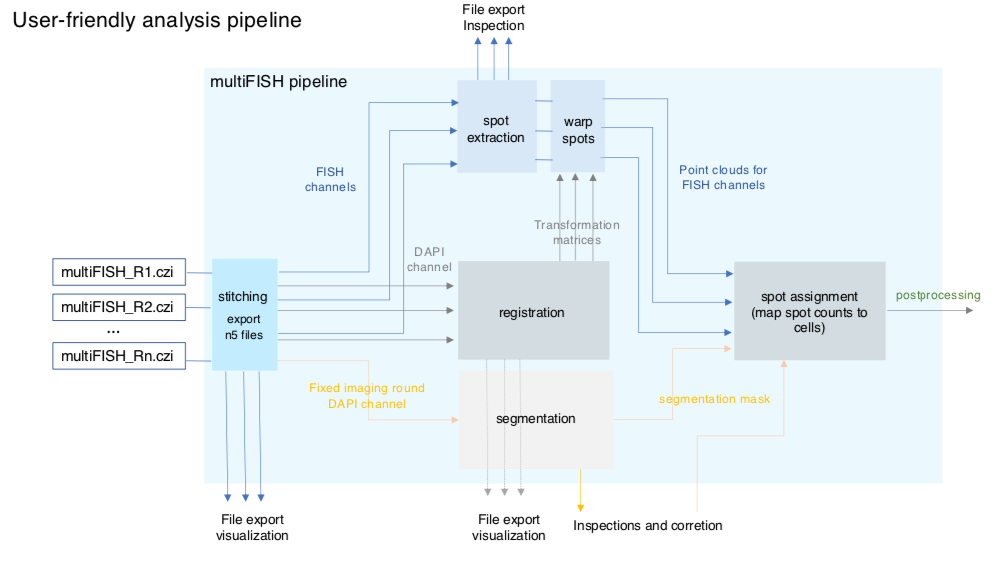
This pipeline analyzes imagery collected using EASI-FISH (Expansion-Assisted Iterative Fluorescence In Situ Hybridization). It includes automated image stitching, distributed multi-round image registration, cell segmentation, and distributed spot detection.
Full documentation is available at https://janeliascicomp.github.io/multifish.
For tech-saavy users, the pipeline can be invoked from the command-line and runs on any workstation or cluster. The only prerequisites for running this pipeline are Nextflow (version 20.10.0 or greater) and Singularity (version 3.5 or greater). If you are running on an HPC cluster, ask your system administrator to install Singularity on all the cluster nodes. Singularity is a popular HPC containerization tool, so many institutional clusters already support it.
To install Nextflow:
curl -s https://get.nextflow.io | bash
To install Singularity on CentOS Linux:
sudo yum install singularity
Clone this repository with the following command:
git clone https://github.com/JaneliaSciComp/multifish.git
Before running the pipeline for the first time, run setup to pull in external dependencies:
./setup.sh
You can now launch the pipeline using:
./main.nf [arguments]