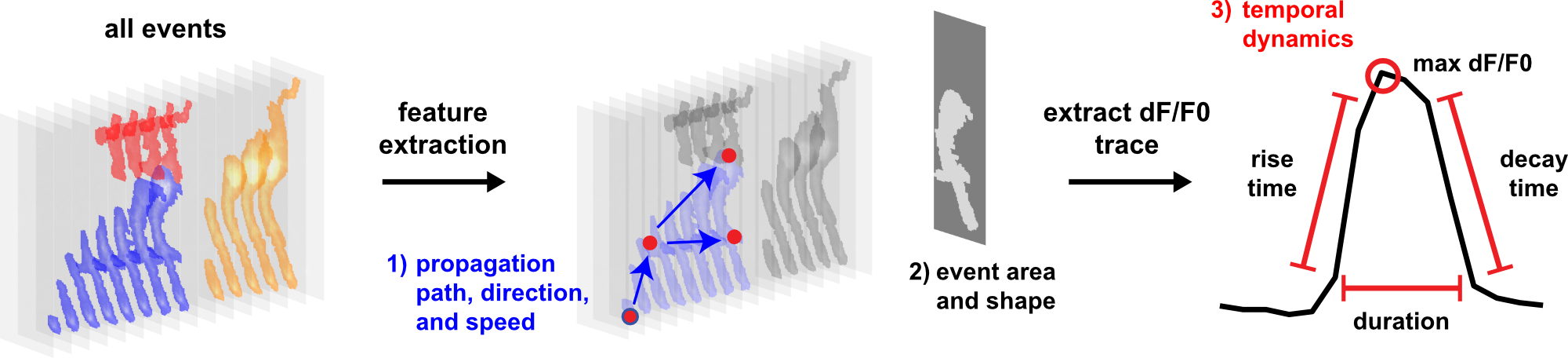
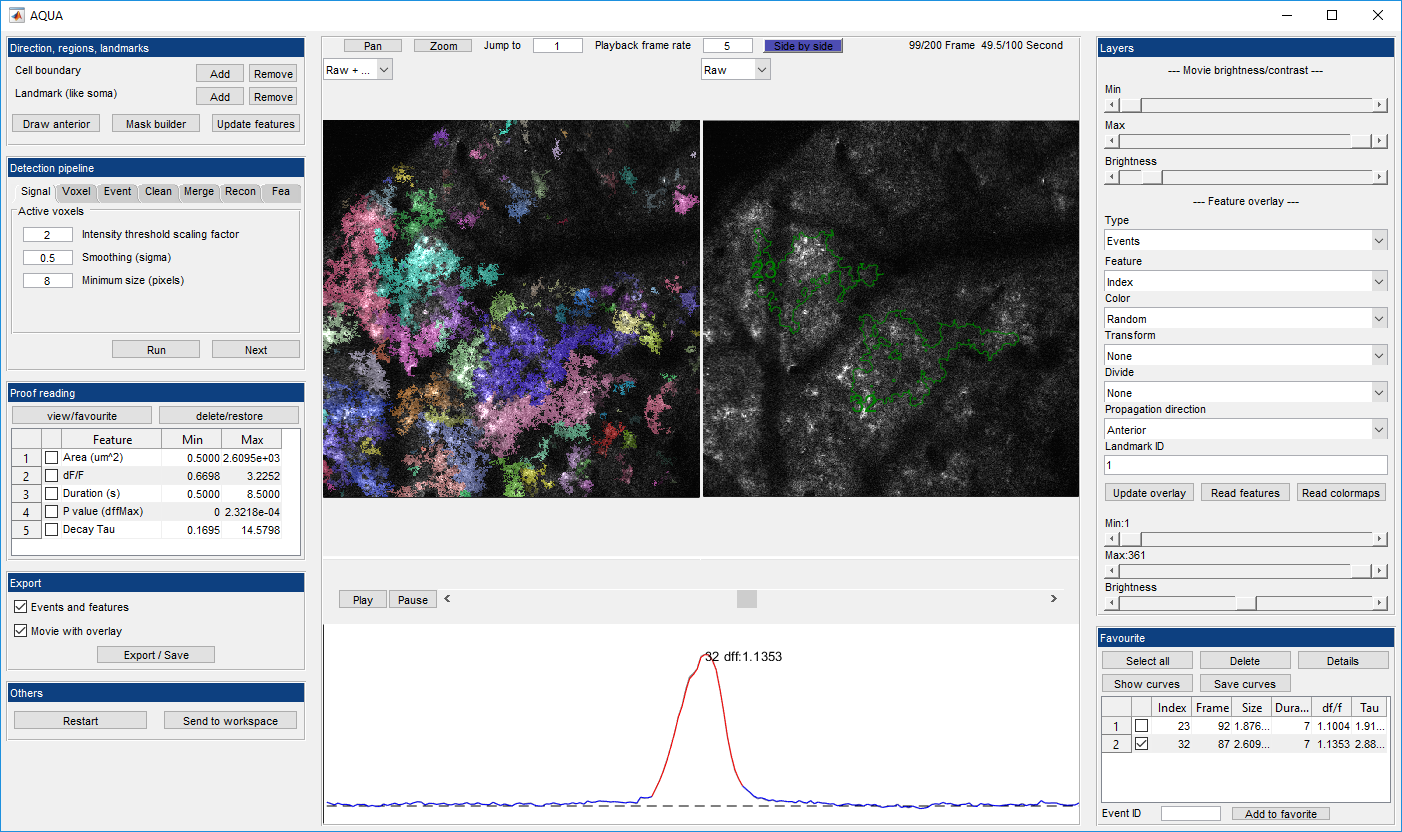
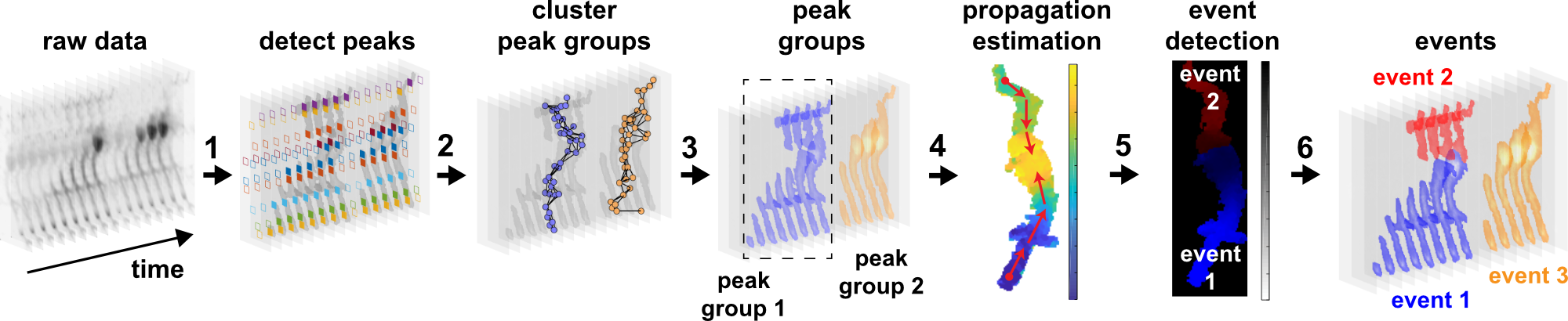AQuA (Astrocyte Quantification and Analysis) is a tool to detect events from microscopic time-lapse imaging data of astrocytes. The algorithm is data-driven and based on machine learning principles, so, potentially, it can be applied across model organisms, fluorescent indicators, experimental modes, cell types, and imaging resolutions and speeds.
- In vivo and ex vivo
- GCaMP, GluSnFr
- And more
- Size and location
- Duration, delta F/F, rising/falling time, decay time constant
- Propagation direction, speed
- And more
- Step by step guide
- Event viewer
- Feature visualizer
- Proofreading and filtering
- Side by side view
- Region and landmark tool
- And more
- Download latest version here.
- Unzip the downloaded file.
- Start MATLAB.
- Switch the current folder to AQuA's folder.
- Double click
aqua_gui.m, or typeaqua_guiin MATLAB command line.
We tested on MATLAB versions later than 2017a. Earlier versions are not supported.
- Download here.
- Put the downloaded
Aqua.jarto the plugins folder of Fiji. - Open Fiji.
- In the
Pluginsmenu, clickAqua. - Open movie and choose project path in AQuA GUI.
Some browsers may show a warning when downloading the 'jar' file. Please choose 'keep file'.
If you are using AQuA for the first time, please read the step by step user guide.
Or you can check the details on output files, extracted features, and parameter settings.
You can try these real data sets in AQuA. These data sets are used in the supplemental of the paper.
We also provide some synthetic data sets. These are used in the simulation part of the paper.
Yizhi Wang, Nicole V. DelRosso, Trisha Vaidyanathan, Michael Reitman, Michelle K. Cahill, Xuelong Mi, Guoqiang Yu, Kira E. Poskanzer, An event-based paradigm for analyzing fluorescent astrocyte activity uncovers novel single-cell and population-level physiology, BioRxiv 504217; doi: https://doi.org/10.1101/504217. [Link to BioRxiv]