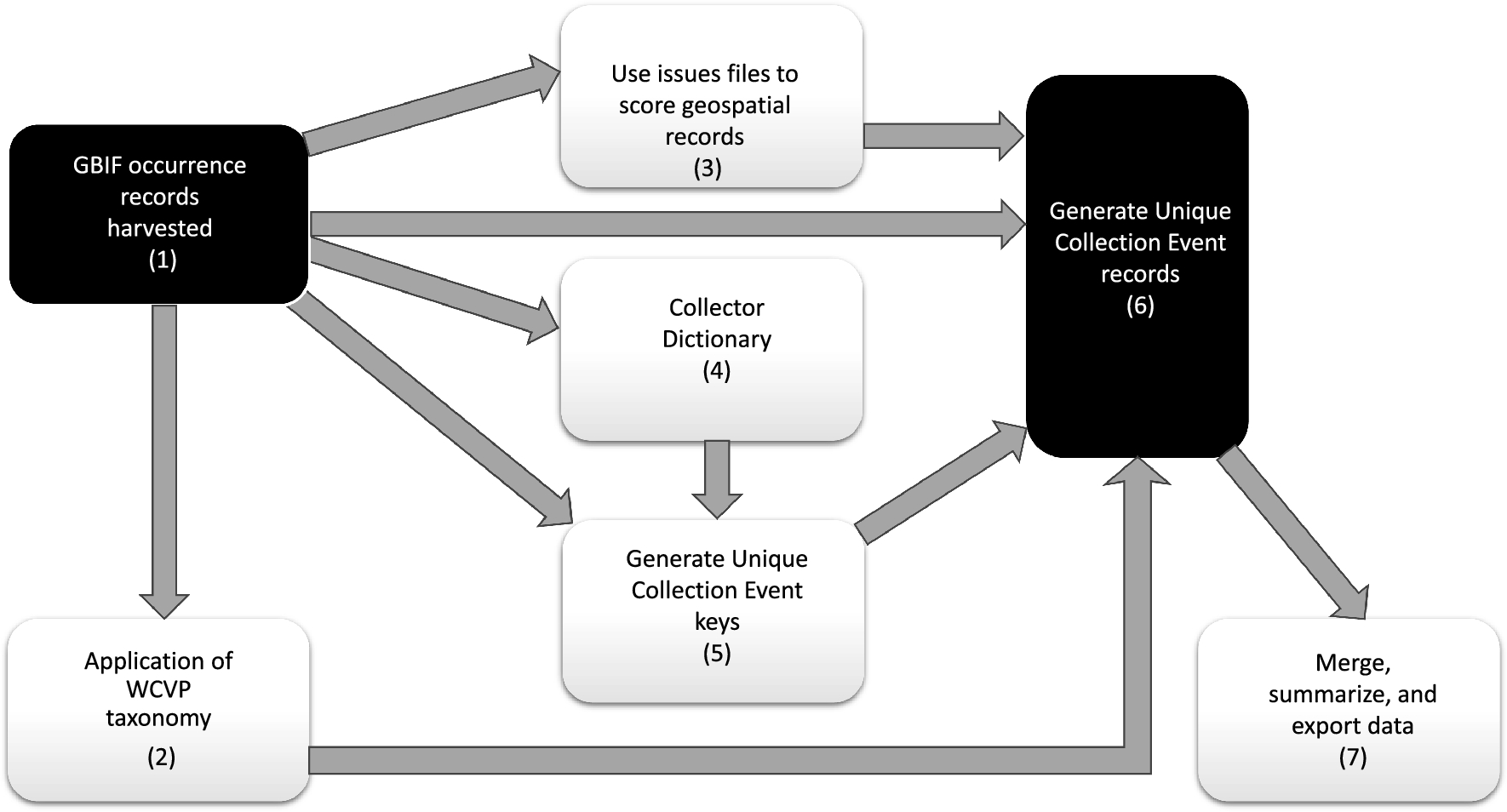
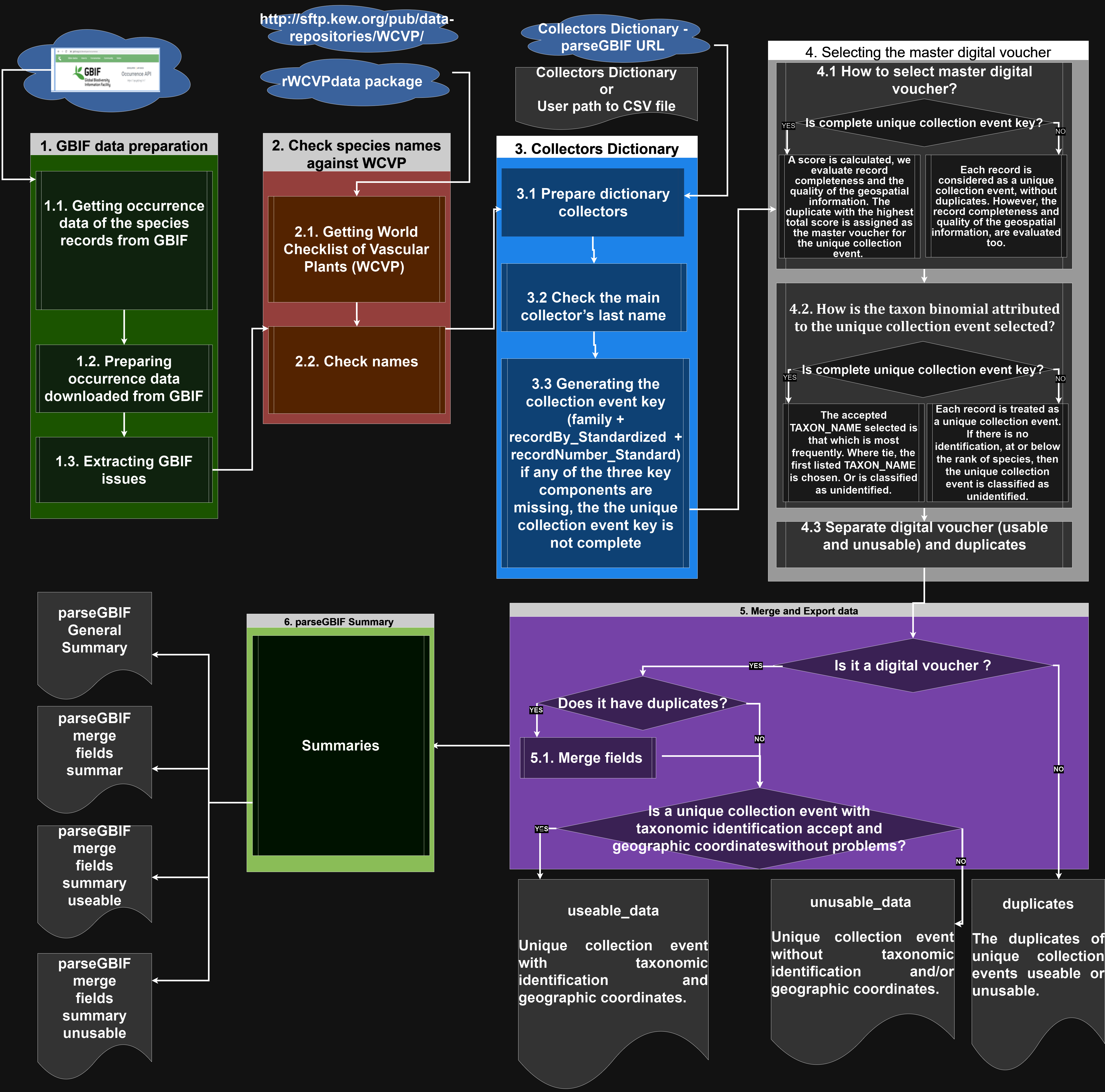
The parseGBIF package is designed to repackage Global Biodiversity Information Facility - GBIF species occurrence records into a format that optimises its use in further analyses. Currently occurrence records in GBIF can include several duplicate digital records, and in the case of vascular plants, for several physical duplicates of unique collection events (biological collections). parseGBIF aims to parse these records to a single, synthetic, record corresponding to a unique collection event to which a standardized scientific name is associated. It does so by providing tools to verify and standardize species scientific names, score the quality of both the naming of a record and of its associated spatial data, and to use those scores to synthesise and parse duplicate records into unique collection events. This Manual provides a brief introduction to parseGBIF, with more information available from Help pages accessed via the help fuction. We believe that this package will be of particular use for analyses of plant occurrence data.
You can install the development version of parseGBIF from GitHub. To install parseGBIF, run
devtools::install_github("pablopains/parseGBIF",
dependencies = TRUE)A new R package to parse plant species occurrence records into unique collection events efficiently reduces data redundancy DOI:10.1038/s41598-024-56158-3
We recommend using the application locally.
parseGBIF::parseGBIF_app()For cloud computing we recommend opening the Jupyter notebook r providing the URL parseGBIF_workflow.ipynb and running the workflow
Consult the parseGBIF Manual, updated, for a case study with a complete and replicable workflow
Please site parseGBIF as:
print(citation("parseGBIF"), bibtex = FALSE)
#> Para citar o pacote 'parseGBIF' em publicações use:
#>
#> de Melo P, Bystriakova N, Lucas E, Monro A (2024). "A new R package
#> to parse plant species occurrence records into unique collection
#> events efficiently reduces data redundancy." _Sci Rep_, *14*(5450),
#> 1-9. doi:10.1038/s41598-024-56158-3
#> <https://doi.org/10.1038/s41598-024-56158-3>.