Port, improve, and benchmark read-based MGnify analysis pipeline for microbiome data using the Galaxy framework
Explore the docs »
mOTUs · SeqPrep
Report Bug · Request Feature
Table of Contents
From October 16th to January 16th, I embarked on a bachelor's project as part of my Bachelor's degree at the Professorship for Bioinformatics. Supervised by Paul Zierep, my task was to port, improve, and benchmark the read-based MGnify analysis pipeline for microbiome data using the Galaxy framework.
Microbiome data analysis is a complex field, necessitating advanced computational tools and methodologies. My project centered on the MGnify V5 Raw reads Pipeline, a well-regarded pipeline in microbiome data analysis.
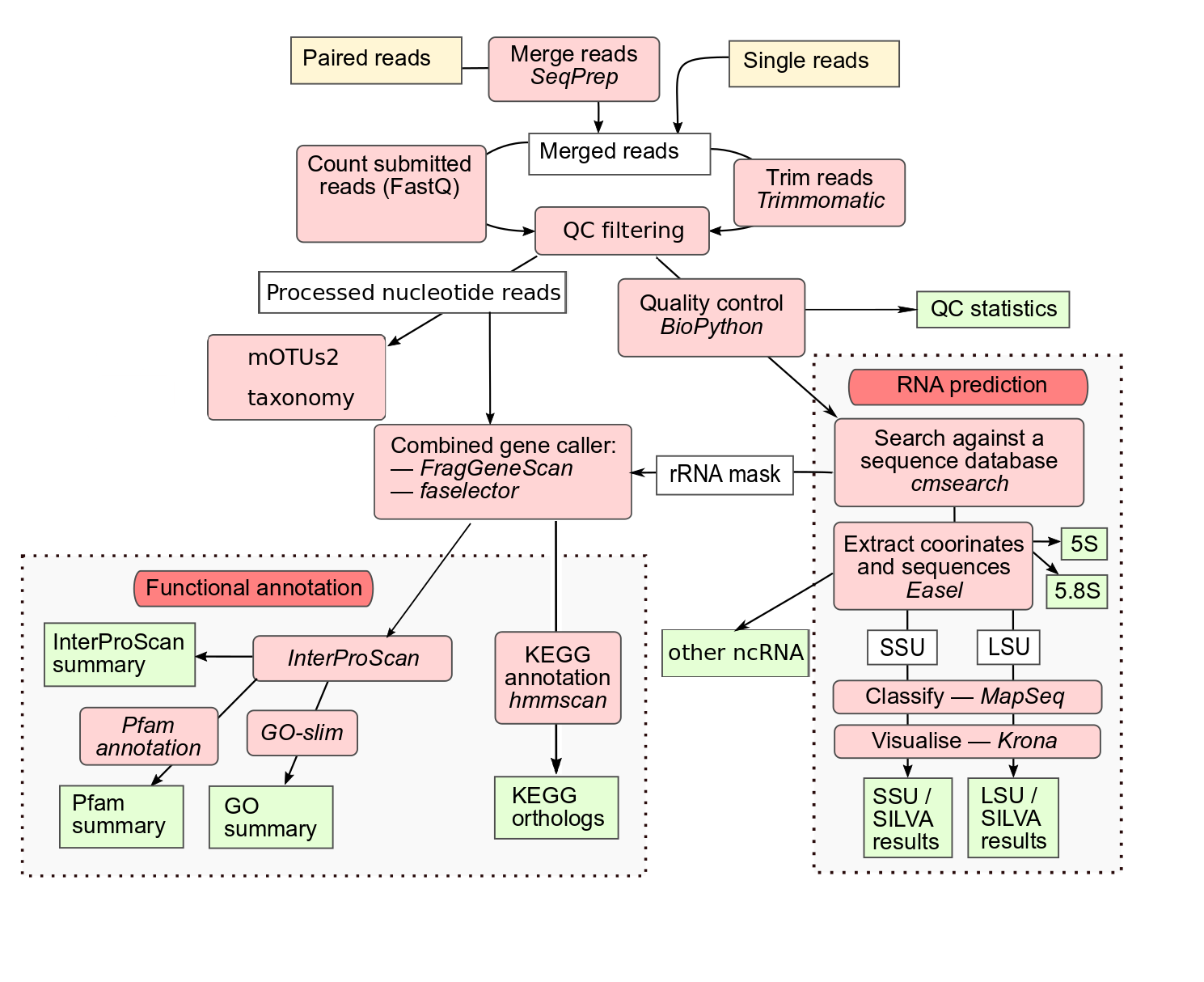
A significant part of my project involved wrapping the tools SeqPrep and mOTUs3 (an updated version compared to mOTUs2 as illustrated in the image) for integration into the Galaxy platform. This integration was crucial as it allows the full workflow of the MGnify V5 pipeline to be executed within Galaxy, making it accessible for a broader range of researchers and facilitating ease of use.
Follow these steps to use SeqPrep on the Galaxy platform and prepare for the upcoming integration of mOTUs.
Before starting, ensure you have created an account on Galaxy. Galaxy offers a web-based platform with a wide array of bioinformatics tools for research.
SeqPrep is going to be integrated into Galaxy and can be used for preprocessing microbiome sequencing data. Here's how to use it:
- Create a Galaxy Account:
- Visit usegalaxy.eu and create an account if you don’t already have one.
- Find the SeqPrep Tool:
- Once logged in to Galaxy, search for "SeqPrep" in the tool search bar.
- Upload Data:
- Upload your microbiome sequencing data to Galaxy.
- Configure SeqPrep:
- Select SeqPrep from the list of tools and configure it according to your specific requirements for your data.
- Run SeqPrep:
- Execute the tool and wait for the results. SeqPrep will process your data
appropriately, preparing it for further analysis.
Currently, mOTUs is not yet directly integrated into Galaxy, but work is underway to make this tool available soon. Once mOTUs is integrated into Galaxy, it will enable efficient and user-friendly metagenomic analyses. Keep an eye on updates in our repository to stay informed about the integration of mOTUs.
The tools and methodologies developed in this project can be applied in various ways to facilitate and enhance microbiome data analysis. Below are some examples of how these tools can be utilized within the Galaxy framework:
-
Microbiome Data Preprocessing with SeqPrep:
- Utilize SeqPrep for the preprocessing of microbiome sequencing data. This includes steps like merging paired-end reads and quality filtering.
- Example Workflow:
- After uploading your paired-end sequencing data to Galaxy, select SeqPrep from the tool menu.
- Configure the tool parameters: specify the input files for forward and reverse reads, and set the merging and quality filtering parameters according to your experiment's requirements.
- Execute the tool to process the data. The output will be a single, merged file of high-quality reads, ready for further analysis.
-
Metagenomic Analysis with mOTUs:
- Deploy mOTUs for metagenomic profiling. This tool helps in identifying and quantifying microbial taxa in the sample data.
- Example Workflow:
- Start with high-quality, preprocessed reads (e.g., output from SeqPrep).
- Select mOTUs from the Galaxy tool menu.
- Input your reads into mOTUs. You may need to specify certain parameters, like the type of analysis (e.g., species-level profiling) or other advanced settings based on your study. Run the tool. mOTUs will output a detailed profile of the microbial community present in your sample.
- This output can be visualized directly in Galaxy using available visualization tools or exported for further analysis.
- Galaxy Environment:
- Galaxy is an open, web-based platform for computational biomedical research. It allows users to reproduce and share analyses.
- Within Galaxy, tools like SeqPrep and mOTUs can be integrated to streamline and simplify the microbiome data analysis pipeline.
- For detailed instructions, use cases, and tutorials on how to use these tools in Galaxy, please refer to the Galaxy Training Network.
Planemo
- Install and Configure Planemo
- Setup Planemo environment on the system.
- Planemo Tutorial
- Complete introductory tutorials for understanding Planemo's functionalities.
- Planemo Documentation
- Extensive reading of the Planemo Documentation to deepen understanding.
SeqPrep
- Understanding SeqPrep
- Research SeqPrep tool and its mechanisms.
- Blueprint for SeqPrep
- Develop a plan for wrapping SeqPrep using Planemo.
- Implement SeqPrep in Galaxy
- Successfully wrap SeqPrep and demonstrate its functionality on Galaxy.
- Contribute to IUC
- Prepare and submit a pull-request to integrate SeqPrep into the IUC main branch. (Here)[galaxyproject/tools-iuc#5719]
mOTUs
-
Explore mOTUs
- Learn about the mOTUs tool and its applications.
-
Plan mOTUs Integration
- Design a strategy for incorporating mOTUs into Galaxy.
-
Galaxy Implementation
- Execute the integration of mOTUs into the Galaxy platform.
-
Testing and Validation
- Ensure mOTUs works seamlessly within Galaxy; perform tests.
-
Contribute to bgruening-galaxytools
- Prepare and submit a pull-request to integrate mOTUs into the galaxytools branch from bgruening main branch.
-
Understand Data Manager Requirements
- Study the specific needs for a data manager for mOTUs.
-
Development Plan
- Draft a comprehensive plan for developing the mOTUs data manager.
-
Implementation
- Develop and integrate the data manager into the Galaxy ecosystem.
-
Optimization and Review
- Optimize for performance and user-friendliness; prepare for peer review.
For a full list of proposed features and known issues, see the open issues.
-
Install Planemo:
- For more information, visit the Planemo documentation.
-
Running Tests:
- To test your Galaxy tools, use the following Planemo command:
planemo test --galaxy_root /path/to/galaxy_root my_tool.xml
- To test your Galaxy tools, use the following Planemo command:
-
Running a Local Galaxy Server:
-
To serve your tools in a local Galaxy instance, use this command:
planemo serve --galaxy_root /path/to/galaxy_root my_tool.xml
-
This will start a local Galaxy server where you can test your tools in a real Galaxy environment.
-
Special thanks to:
- Paul Zierep: For his mentorship and expertise in guiding the development and implementation of the project, particularly in the areas of bioinformatics and data analysis.
- Rand: For the significant assistance with Planemo and SeqPrep, facilitating key aspects of the project's development.
- Galaxy Project Team: For providing an excellent platform for bioinformatics research.
- SeqPrep and mOTUs Developers: For their contributions to microbiome data analysis tools.