Open source comparisons of multiple different prophage predictions
There are multiple different ways of identifying prophages in bacterial genomes, and this is an open source way of comparing them. Please feel free to clone this repo, add your tool or code, and then make a pull request.
Prophages are viruses that are integrated into bacterial genomes. A few computatational biologists are keen to identify those specific regions, because they are more interesting than the rest of the genome. For more about prophages, take a look at the home pages for some of the tools listed here.
This site is not intended to be a gentle introduction to prophages, but a FAIR (findable, accessible, interoperable, and reusable) data resource for comparing prophage prediction software.
To run the tests, first clone the repository and pull the files (requires git and git lfs)
git clone https://github.com/linsalrob/ProphagePredictionComparisons.git
cd ProphagePredictionComparisons
git submodule update
git lfs install
git lfs updateThen run the pipelines (requires snakemake and conda)
snakemake -s snakefiles/virsorter.snakefile --use-conda # --profile slurm or -j 16 etc...If you develop prophage prediction software, clone the repository and implement your tool using a snakemake pipeline. There are several examples in the snakefiles directory. We have also defined conda environments for each of the tools (see the note below).
Once your tool is working, use it to predict the prophages in the genbank folder, and use the scripts to calculate true positive, true negative, false positive, false negative and related statistics.
The jupyter notebooks can be used to plot your data and make images like those below.
If you go to all that work, please make a pull request and we will update this site with your code.
Note: The conda environments are not yet working reliably. Please let us know if you have issues.
We have:
- Phage Finder (original citation) Version used: v2.1
- PhiSpy (original citation) Version used: 4.2.6
- VirSorter (original citation) Version used: v1.0.6
- Phigaro (original citation) Version used: v2.3.0
- DBSCAN-SWA (original citation) Version used: 2e61b95
- PhageBoost (original citation) Version used: 0.1.7
- Virsorter2 (original citation) Version used: 2.2.1
We could not install:
- LysoPhD - We can not find this available online anywhere
- ProphET - This requires legacy BLAST and EMBOSS packages and we could not get it to install and run.
If you know of other tools that should be included please let us know or make a PR.
We manually curated the prophages in the bacterial genomes in the genbank files. For each phage we mark both
the prophage region, and we mark each prophage gene as being a phage gene with a unique is_phage tag. We run the
prediction software on those genbank files, and then compare the predictions with our manual curations.
We need more manually curated genomes! Please contribute by adding more manually curated genomes to our data set.
Our dataset of manually curated genomes is a start, and we welcome submissions from anyone. To add a new genome:
- Please generate a GenBank format file with the complete bacterial genome
- For the
CDSentries that are phages, please add the flag/is_phage="1"to the entry (the value doesn't matter, we check for the presence of theis_phageflag and that the value is not zero) - Make a clone of this repository and add your genome(s)
- Make a pull request to add your genome(s) from your clone to the master branch
We welcome annotated microbial genomes from all sources, but we ask that you please manually curate the presence of phage, because it is that gold-standard manual curation that allows us to accurately compare tools.
Since we have a notion of truth, we calculate and plot:
- true positives (TP)
- true negatives (TN)
- false positives (FP)
- false negatives (FN)
- accuracy : the ratio of the correctly labeled phage genes to the whole pool of genes
- precision : the ratio of correctly labeled phage genes to all predictions
- recall : the fraction of actual phage genes we got right
- specficity : the fraction of non phage genes we got right
- F1 score : the harmonic mean of precision and recall, and is the best measure when, as in this case, there is a big difference between the number of phage and non-phage genes
Note that these plots are generated by the jupyter notebooks we provide, but please repeat them and let us know if we made an error!
We plotted the accuracy, precision, recall, and specificity of the different callers, and in this plot each subplot has the same axis.
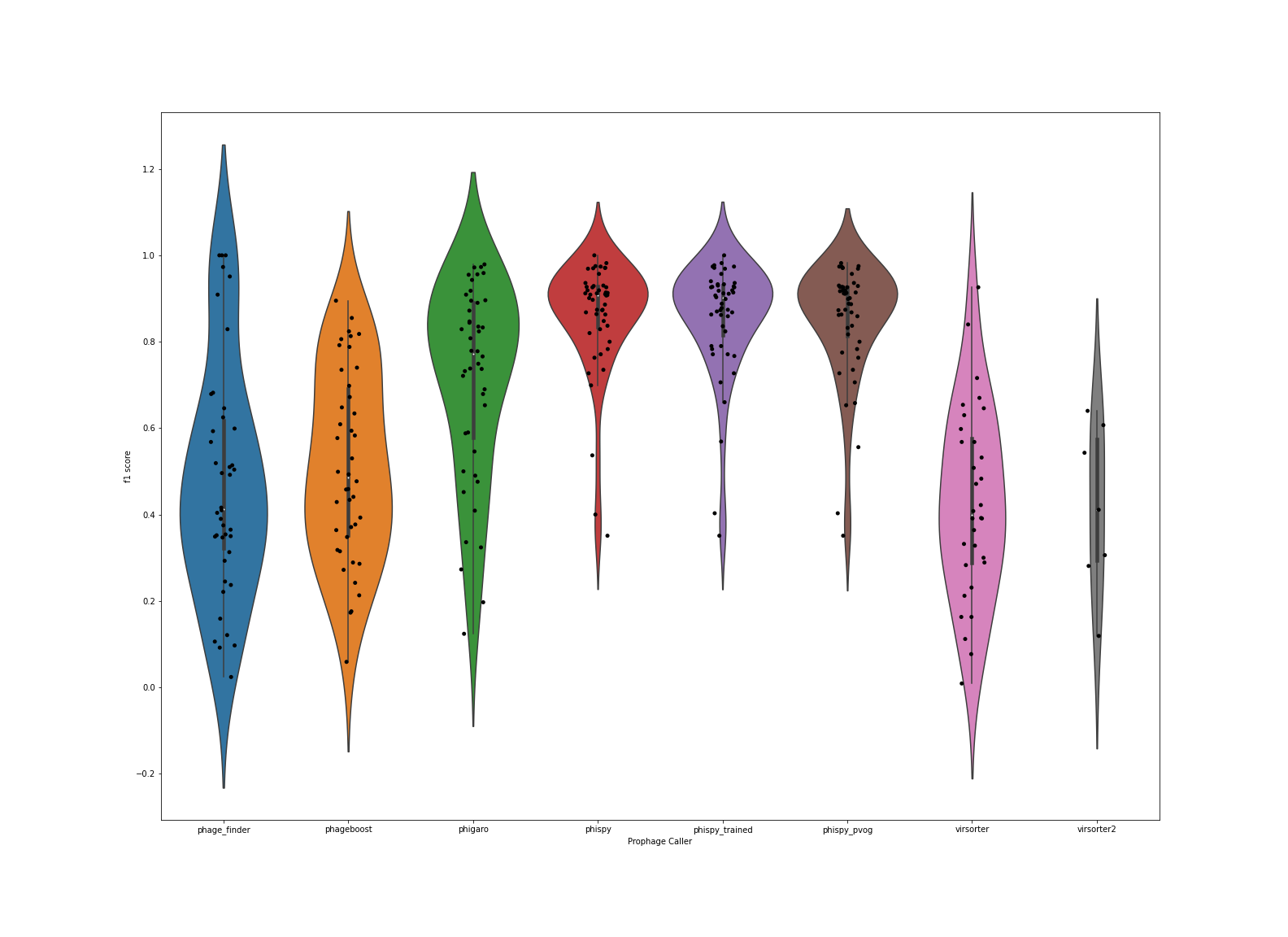
As noted above, however, these are probably not the most robust data since we have a lot of non-phage genes (ie. everything in the genome that is not a prophage), and only a relatively few phage genes. So we rely more on F1 score as shown here:
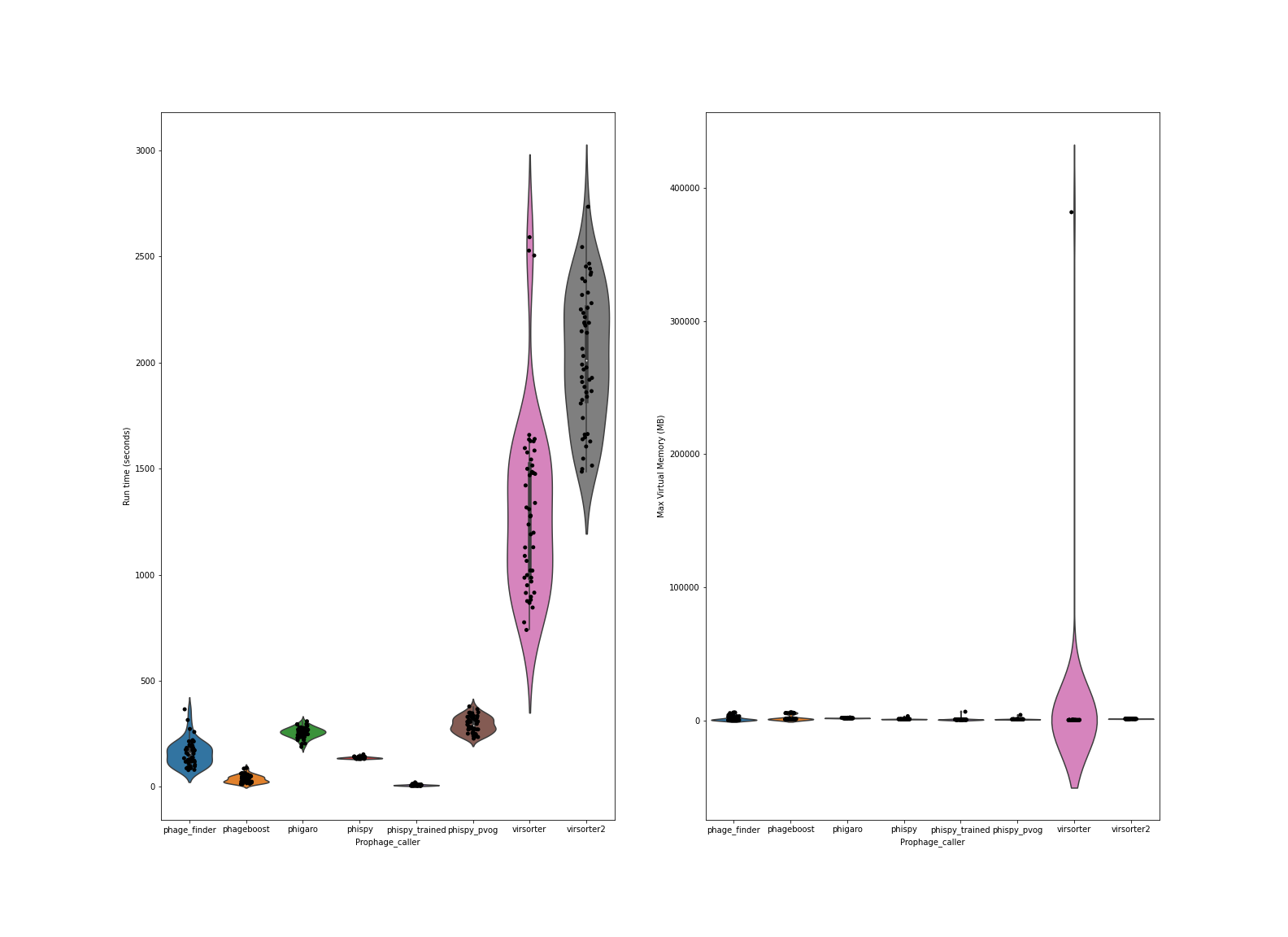
Speed is of the essence, and this is where each of the prophage callers really begin to differ. This plot shows time (seconds) to complete the predictions, and amount of memory consumed.
Not much! You should always take benchmarks with a grain of salt, because whoever made them (see below) usually has a vested interest int their outcome.
You should note, however, that phage_finder, the OG of prophage identification is still one of the most robust methods.
This site was put together by Rob Edwards to compare prophage predictions. Help him out with curated genomes!