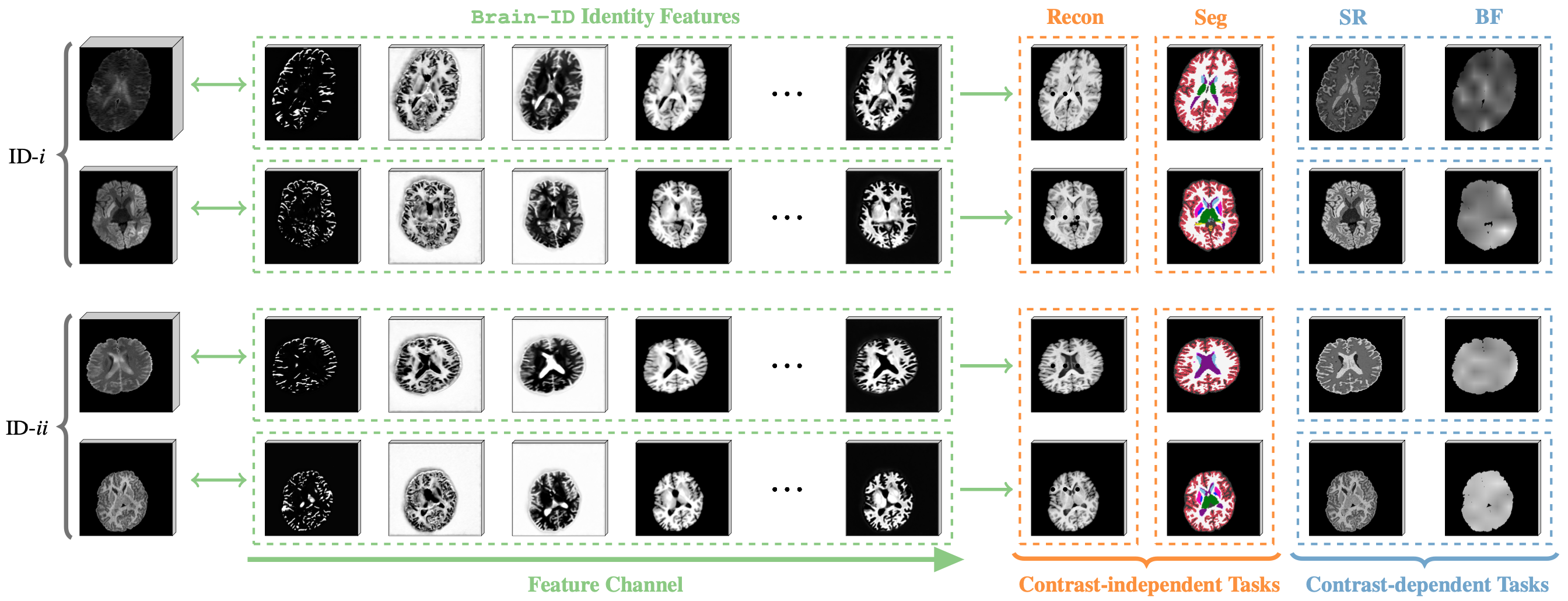
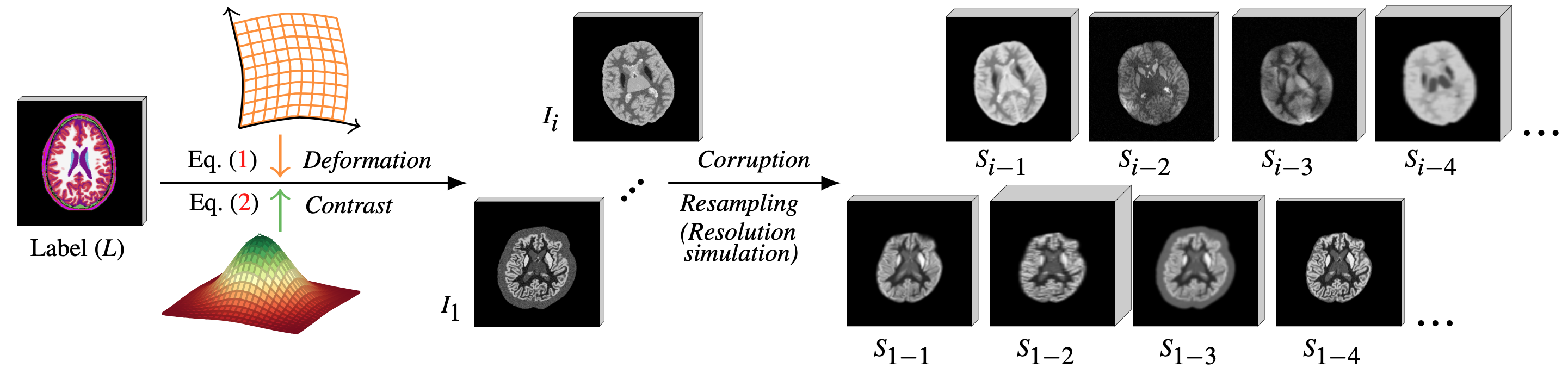
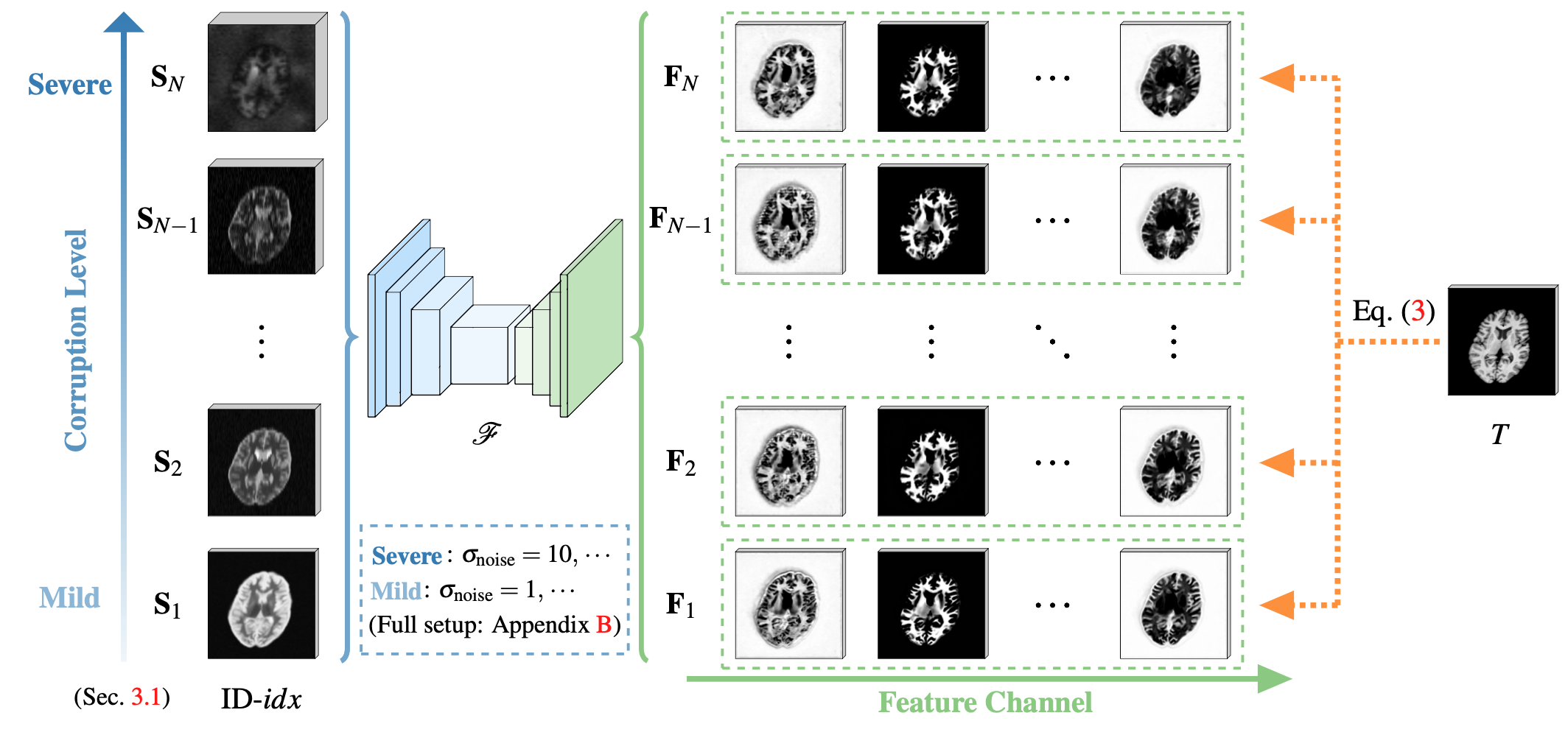
Peirong Liu1, Oula Puonti1, Xiaoling Hu1, Daniel C. Alexander2, Juan Eugenio Iglesias1,2,3
1Harvard Medical School and Massachusetts General Hospital
2University College London 3Massachusetts Institute of Technology
[06/17/2024] Check out our newest MICCAI2024 work here on a contrastic-agnosic model for images with abnormalities with pathology-encoded modeling.
Training and evaluation environment: Python 3.11.4, PyTorch 2.0.1, CUDA 12.2. Run the following command to install required packages.
conda create -n brainid python=3.11
conda activate brainid
git clone https://github.com/peirong26/Brain-ID
cd /path/to/brain-id
pip install -r requirements.txt
cd /path/to/brain-id
python scripts/demo_synth.py
You could customize your own data generator in cfgs/demo_synth.yaml.
Please download Brain-ID pre-trained weights (brain_id_pretrained.pth), and test images (T1w.nii.gz, FLAIR.nii.gz) in this Google Drive folder, and move them into the './assets' folder.
Obtain Brain-ID synthesized MP-RAGE & features using the following code:
import os, torch
from utils.demo_utils import prepare_image, get_feature
from utils.misc import viewVolume, make_dir
img_path = 'assets/T1w.nii.gz' # Try: assets/T1w.nii.gz, assets/FLAIR.nii.gz
ckp_path = 'assets/brain_id_pretrained.pth'
im, aff = prepare_image(img_path, device = 'cuda:0')
outputs = get_feature(im, ckp_path, feature_only = False, device = 'cuda:0')
# Get Brain-ID synthesized MP-RAGE
mprage = outputs['image']
print(mprage.size()) # (1, 1, h, w, d)
viewVolume(mprage, aff, names = ['out_mprage_from_%s' % os.path.basename(img_path).split('.nii.gz')[0]], save_dir = make_dir('outs'))
# Get Brain-ID features
feats = outputs['feat'][-1]
print(feats.size()) # (1, 64, h, w, d)
# Uncomment the following if you want to save the features
# NOTE: feature size could be large
#num_plot_feats = 1 # 64 features in total from the last layer
#for i in range(num_plot_feats):
# viewVolume(feats[:, i], aff, names = ['feat-%d' % (i+1)], save_dir = make_dir('outs/feats-%s' % os.path.basename(img_path).split('.nii.gz')[0]))You could also customize your own paths in scripts/demo_brainid.py.
cd /path/to/brain-id
python scripts/demo_brainid.py
Use the following code to train a feature representation model on synthetic data:
cd /path/to/brain-id
python scripts/train.py anat.yaml
We also support Slurm submission:
cd /path/to/brain-id
sbatch scripts/train.sh
You could customize your anatomy supervision by changing the configure file cfgs/train/anat.yaml. We provide two other anatomy supervision choices in cfgs/train/seg.yaml and cfgs/train/anat_seg.yaml.
Use the following code to fine-tune a task-specific model on real data, using Brain-ID pre-trained weights:
cd /path/to/brain-id
python scripts/eval.py task_recon.yaml
We also support Slurm submission:
cd /path/to/brain-id
sbatch scripts/eval.sh
The argument task_recon.yaml configures the task (anatomy reconstruction) we are evaluating. We provide other task-specific configure files in cfgs/train/task_seg.yaml (brain segmentation), cfgs/train/anat_sr.yaml (image super-resolution), and cfgs/train/anat_bf.yaml (bias field estimation). You could customize your own task by creating your own .yaml file.
-
Brain-ID pre-trained weights and test images: Google Drive
-
ADNI, ADNI3 and AIBL datasets: Request data from official website.
-
ADHD200 dataset: Request data from official website.
-
HCP dataset: Request data from official website.
-
OASIS3 dataset Request data from official website.
-
Segmentation labels for data simulation: To train a Brain-ID feature representation model of your own from scratch, one needs the segmentation labels for synthetic image simulation and their corresponding MP-RAGE (T1w) images for anatomy supervision. Please refer to the pre-processing steps in
preprocess/generation_labels.py.
For obtaining the anatomy generation labels, we rely on FreeSurfer. Please install FreeSurfer first and activate the environment before running preprocess/generation_labels.py.
After downloading the datasets needed, structure the data as follows, and set up your dataset paths in BrainID/datasets/__init__.py.
/path/to/dataset/
T1/
subject_name.nii
...
T2/
subject_name.nii
...
FLAIR/
subject_name.nii
...
CT/
subject_name.nii
...
or_any_other_modality_you_have/
subject_name.nii
...
label_maps_segmentation/
subject_name.nii
...
label_maps_generation/
subject_name.nii
...
@InProceedings{Liu_2023_BrainID,
author = {Liu, Peirong and Puonti, Oula and Hu, Xiaoling and Alexander, Daniel C. and Iglesias, Juan E.},
title = {Brain-ID: Learning Contrast-agnostic Anatomical Representations for Brain Imaging},
booktitle = {European Conference on Computer Vision (ECCV)},
year = {2024},
}