CellDive (src/) is a tool for exploring clonality of lymphocytes and differential expression based on single-cell RNA-seq:
- clonality rules specification based on the alpha and beta chains of T-cell and B-cell receptors (TCR and BCR)
- clustering into
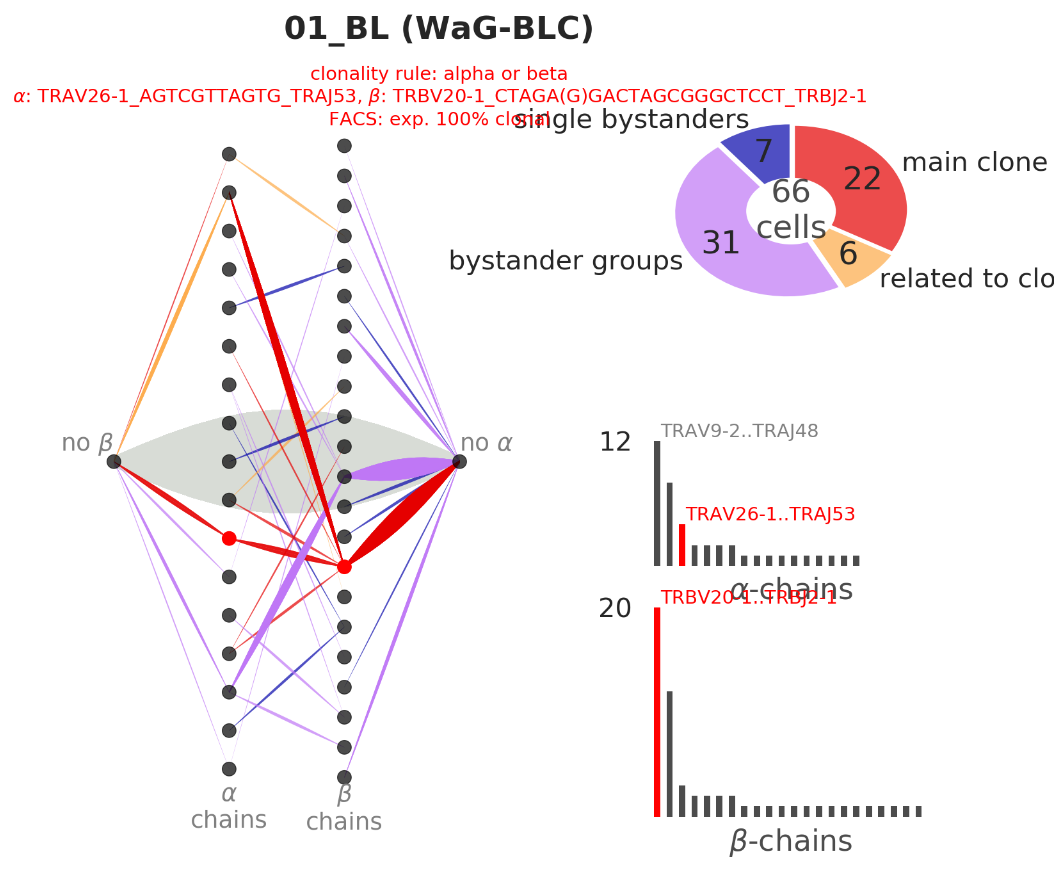
main-clone,related-to-main-clone,bystander groupsandsingle bystandersbased on the clonal rules - bipartite graph visualization of the full clonality information from the samples
- differential expression (DE) between specified clones within the same sample (no batch effects!)
- gene sets overview accross samples and DE methods
CellDive has been applied (example/) to a dozen of human cutaneous lymphoma samples. The clonality rules were used to partition each sample into a malignant clone (main-clone) and healthy cells (bystanders). Differential expression analyses was applied to the two clusters (no batch effect).
Pesho Ivanov (advisor Martin Vechev), Software Reliability Lab, ETH Zurich:
- code and execution
- alpha and beta chain language rules for specifying clones. each cell is colored depending on the clone
- bipartite graph with left nodes for alpha-chains, right nodes for beta-chains, and cells as edges (possibly connected to No-alpha and No-beta nodes)
Data used to produce example/, Department of Dermatology, University Hospital Zurich:
- Yun-Tsan, Desislava Ignatova, Emmanuella Guenova
Note: The code has been written in 2017--2018 so it may be outdated.