If you use trfermikit, please cite the paper: https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btab805/6448210
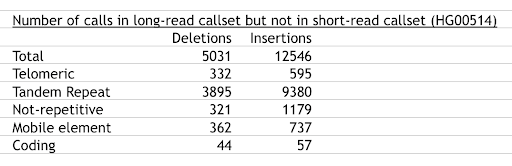
Most SVs missed by short-read callers lie in tandem repeats:
(based upon Supplementary Data 53 of Chaisson et al 2019 and private communication with Mark Chaisson). This observation prompted us to optimize existing SV callers on tandem repeats.
As callers exist to capture SVs in tandem repeats where the repeat unit is smaller than 6bps, known in the community as Short Tandem Repeats (STRs), we designed trfermikit to pick up SVs that manta missed in tandem repeats with repeat units larger than 6bps, known as Variable Number Tandem Repeats (VNTRs).
We assessed the performance of trfermikit and manta, in both cases relative to a long-read benchmark callset, on VNTRs. We found that for DELs: (a) trfermikit has better sensitivity-FDR trade-offs than manta and (b) trfermikit is complementary to manta. In the figures, the red circle indicates the default operating point of trfermikit.
When considering INSs, trfermikit: (a) has similar sensitivity-FDR trade-offs to manta and (b) does not significantly complement manta.
Thus trfermikit is a tool to discover DELs missed by manta in VNTRs.
trfermikit is based upon the fermikit pipeline for deep Illumina resequencing data, which assembles reads into unitigs, maps them to the reference genome, and then calls variants from the alignment.
trfermikit biases the minimap2 alignment step of the fermikit pipeline towards revealing deletions
by:
- increasing the reward for single-base matches
- increasing the penalty for single-base mismatches
- decreasing the gap-open penalties (there are two because the cost function of gap length is piecewise linear)
- decreasing the gap-extension penalties
This recovers a lot of events that a more stringent caller would throw out. The cost is that an elevated number of false discoveries are made. trfermikit mitigates this by:
- throwing out calls that are supported by "dirty" fermikit unitigs (essentially, those that have lots of small blocks when aligned to the reference or those whose mapping quality is zero)
- sparsifying “clusters” of calls
git clone https://github.com/petermchale/trfermikit
cd trfermikit
bash install.sh
conda activate trfermikit
Only installation on Linux x86_64 is currently supported.
Assuming that the path to this directory on your filesystem is
${root}, and that the trfermikit conda environment has been activated, usage is:
PATH="${root}:$PATH"
trfermikit [OPTIONAL_ARGUMENTS] REQUIRED_ARGUMENTS
Required arguments are:
--output STR
STR specifies the path to the directory where the results will be stored.
--reference STR
STR specifies the path to the reference fasta (without the ".fa" suffix).
--alignments STR
STR specifies the path to a set of short-read alignments (without the ".cram" suffix").
The cram index is assumed to be present at the same PATH.
--threads INT
INT specifies the number of threads to be used.
Optional arguments are:
--hg19
Use hg19 build of the human reference genome.
If this flag is not specified, trfermikit uses build hg38.
--functional-regions PATH
Restrict examination to those tandem repeats that lie in the regions indicated by PATH
(without the ".bed.gz" suffix).
[default value: None]
--min-repeat-length INT
Only consider tandem repeats
whose total number of bps is larger than INT
[default value: 100].
Discovered SVs are output in indexed vcf format to the results directory at
fermikit.raw.decomposed.normalized.${svtype}.unitigSupport.thinned.vcf.gz
where ${svtype} is either DEL or INS. As indicated in Impact,
you'll only be interested in the DEL call set.
Tandem-repeat regions used to discover SVs appear in the results directory in indexed bed format at
regions.bed.gz
The parameter configuration used to make the discoveries appear in the results directory at config.json.
By default, the pipeline takes about 2 hours (assuming a 70X genome),
mainly because the search is confined to tandem repeats with total lengths larger than 100bp
(see the min-repeat-length option).
Long-read sequencing data tell us that such tandem repeats harbor most of the
tandem-repeat-associated DELs larger than 50bp.
- Create a docker container and nextflow workflow (with nextflow processes for "make-regions", "make-calls" and "filter-calls") and register both at dockstore.
- Support bams